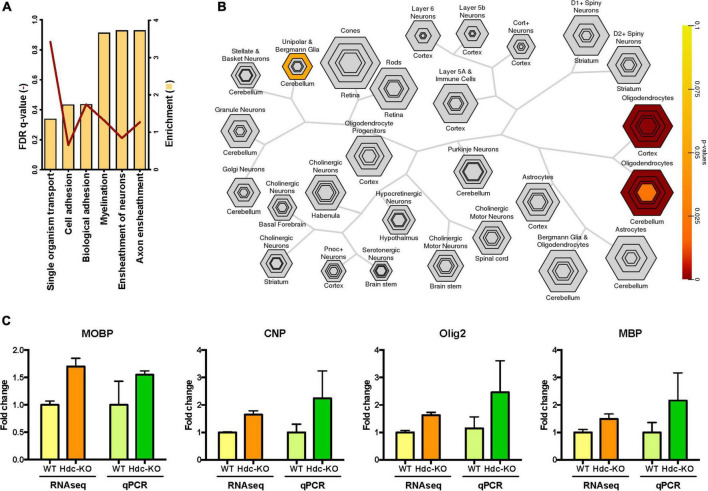
FIGURE 1.
Striatally enriched genes in histidine decarboxylase-knockout (Hdc-KO) mice associated with white matter. (A) Gene ontology analysis (target set n = 778, background set n = 7376) revealed that the most enriched genes in the dorsal striatum (bars) were involved in axon ensheathment, ensheathment of neurons, and myelination. False discovery rate (FDR) corrected q-values are represented as lines. (B) Cell specific expression analysis (CSEA) comparing Hdc-KO with WT in the dorsal striatum (candidate gene list n = 40). Output is organized hierarchically by cell type to recapitulate biological relationships, with size corresponding to the size of the cell population and nested groups representing more specific populations to that cell type. Enriched genes were specific to oligodendrocytes. (C) Parallel changes in gene expression levels of four oligodendrocyte markers in the dorsal striatum in Hdc-KO mice by RNA-Seq and qPCR analysis.