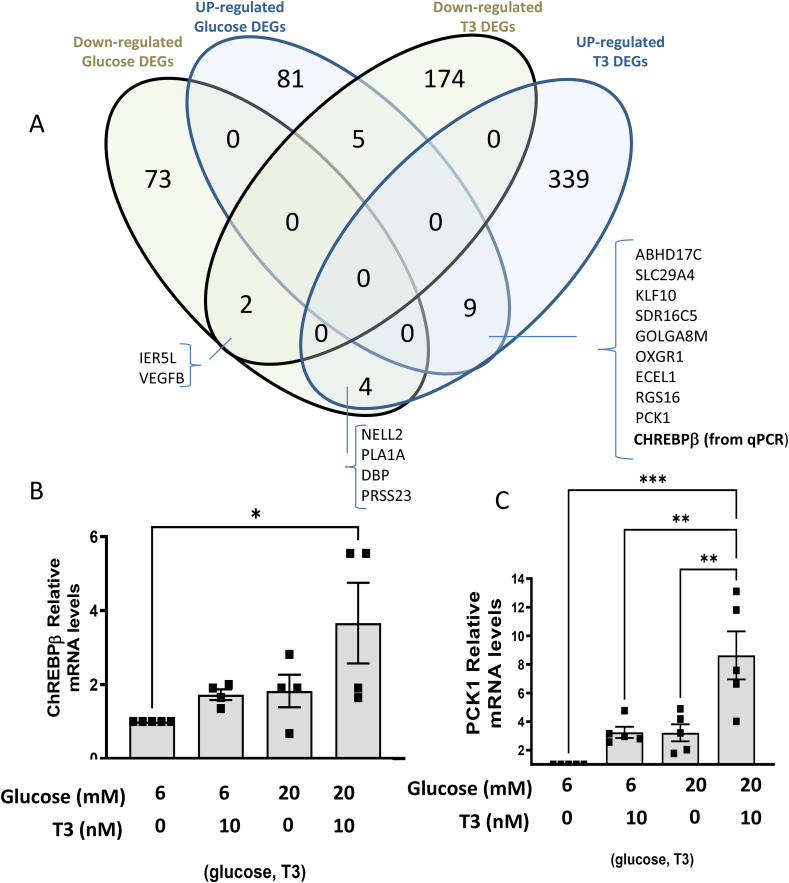
Figure 4.
Determining genes affected by high glucose and T3. A. A Venn diagram showing the up- and down-regulated genes found differentially expressed either following glucose or T3 treatment. Only genes that were found significantly differentially expressed in either condition (at BH Adj P < 0.05 and no logFold change cut-off) were compared. Target validation, from the same donors used for the RNA-Seq, with indicated glucose and T3 concentrations. ChREBPβ and Pck1 mRNA levels were determined by qRT-PCR. Data are the means ± SEM of three independent experiments. All mRNA levels were normalized to β-actin. ∗P < 0.05; ∗∗P < 001; ∗∗∗P < 005 by one-way ANOVA.