Figure 4.
Meta-analysis of COVID-19 patient BALF scRNA-seq reveals differential expression of inflammasome pathway by disease state
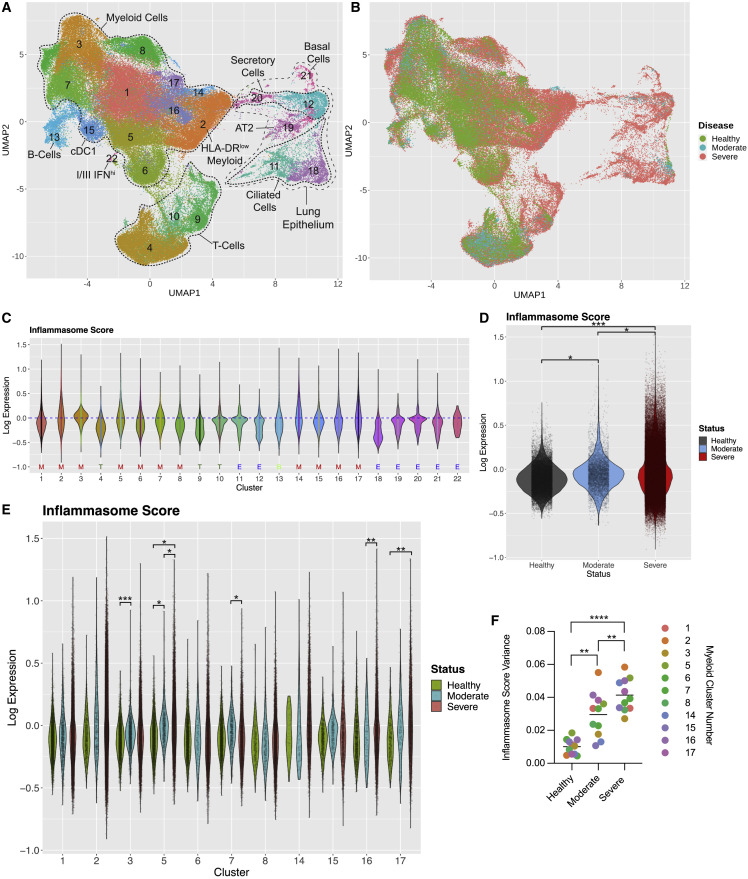
(A) UMAP of healthy and COVID-19 patient BALF scRNA-seq from pooled studies (healthy: n = 3; mild/moderate COVID-19: n = 5; severe COVID-19: n = 26). Each dot represents a cell, and each dot is colored according to cluster identity. Clusters are annotated by cell type. ‡ denotes the HLA-DRlow myeloid population.
(B) UMAP shown in (A) but colored by patient disease state (healthy, mild/moderate, or severe).
(C) Inflammasome scores for each cluster of the UMAP shown in (A). General cluster identity indicated below cluster number: M, myeloid cells; T, T cells; B, B cells, and E, epithelial cells.
(D) Inflammasome scores for all myeloid cells in COVID-19 disease states and healthy patients. Each dot represents a cell.
(E) Inflammasome scores for individual cells (individual points) within each myeloid cluster across COVID-19 disease states.
(F) Inflammasome score variance within each myeloid cluster by disease state. (C)–(F) include projected datasets. Statistical calculations are described in the STAR Methods and text, except for (F) which was determined by paired two-sided t test ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, or ∗∗∗∗p < 0.0001.
See also Figure S4.