Fig. 2. Plasmid persistence shown as the estimated ratio of trfA/16S rRNA genes over a period of eight days in liquid serial batch cultures in the absence of selection.
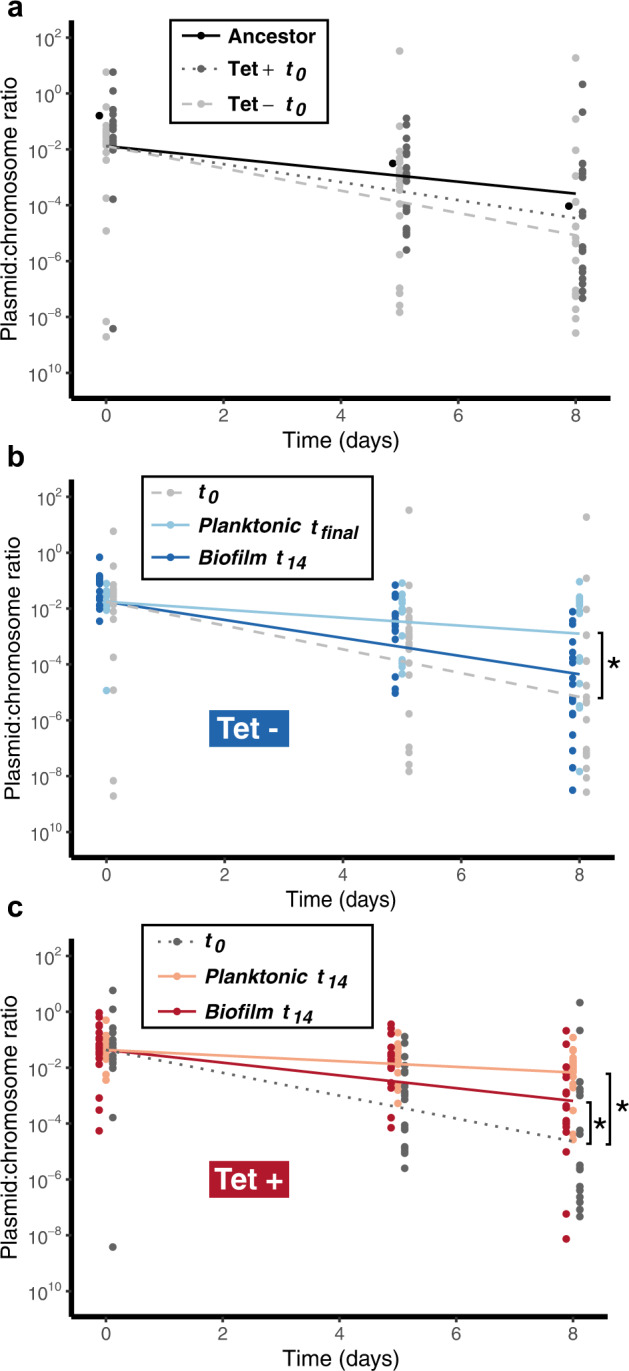
a The ancestor used to inoculate the flow cells at t−4 compared to clones isolated from all the biofilms harvested after four days of growth with tetracycline, t0, showing similar plasmid persistence dynamics (Tet+ and Tet−: populations treated with and without tetracycline after t0); b clones isolated from the Tet− populations, showing a drastic increase in plasmid persistence after evolution in planktonic but not in biofilm populations (note that two of the three planktonic populations no longer contained detectable plasmids after day 6 and 10, respectively, requiring analysis on these days instead of t14—the final sampling times are therefore referred to as tfinal); c clones isolated at t0 and t14 of the Tet+ evolution experiment, showing an increase in plasmid persistence over time for all populations. Lines are the output of the log-linear model for each group. *Denotes a significant difference (t-test p-value < 0.05) between the groups pointed out by the brackets: planktonic, tfinal versus t0, and biofilm t14 versus t0. The plasmid persistence data from t0 in the Tet+ experiment were reported by us previously43.
