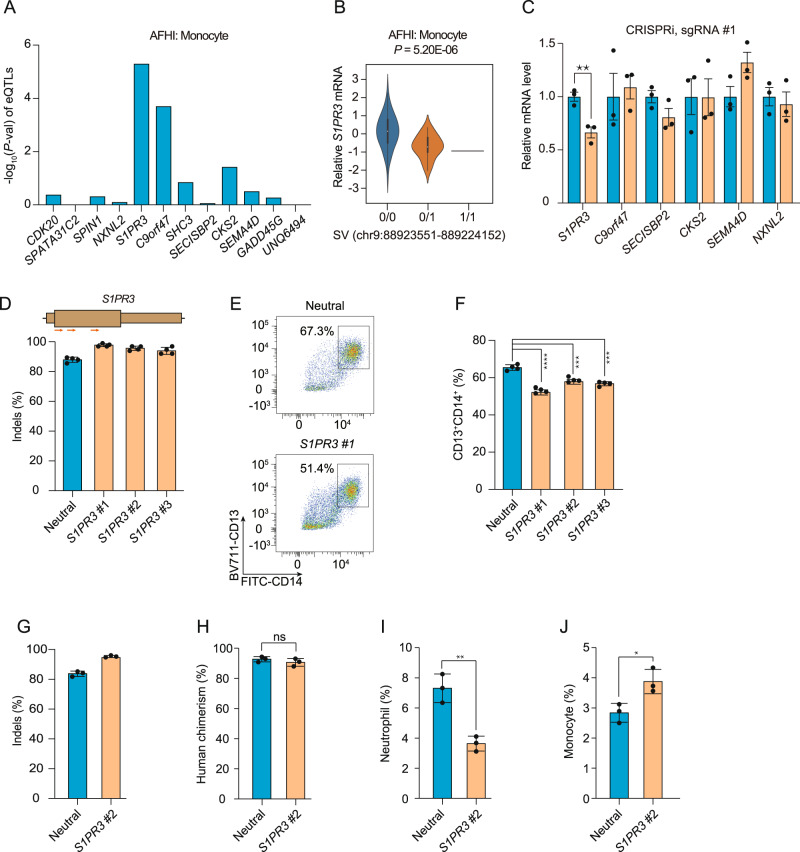
Fig. 2. Genome and epigenome editing implicates S1PR3 in the 9q22.1 monocyte association.
All p-values are derived from two-sided t-tests (unless otherwise indicated) and are not adjusted for multiple comparisons. A eQTL results between the 9q22.1 SV and genes within 1-Mb window in monocytes using data from from MESA, including n = 77 AA and n = 92 Hispanic/Latino participants. AFHI: African American and Hispanic/Latino. B Violin plot (with minima, maxima, median, and inter-quartile range) demonstrating the correlation between the 9q22.1 SV genotype and expression of S1PR3, in n = 169 from MESA. C Expression of genes within a 2 Mb window in THP-1 cells expressing dCas9-KRAB after transduction with an sgRNAs targeting the SV (orange) as compared to a neutral locus control sgRNA (blue). Relative mRNA level of each gene was represented by mean ± standard deviation (SD). N = 3 biological replicates, where each replicate is a unique cellular transduction by sgRNA cassette. **P = 0.009. Location of sgRNAs designed for CRISPRi are indicated in Fig. S8C. D–F S1PR3 gene editing impaired monocyte differentiation in vitro. D Editing efficiency in HSPCs following 3xNLS-SpCas9:sgRNA electroporation with the indicated sgRNA. Gene edits were measured after 4 days of electroporation (N = 4 biological replicates). Location of S1PR3 coding sequence targeting sgRNAs are indicated above. E Representative flow cytometry indicating CD13 + CD14 + cell populations from the neutral locus and S1PR3 targeting group after 12-day differentiation. F CD13 + CD14 + percentage in the S1PR3 targeting group and the neutral locus targeting group. N = 4 replicates where each replicate is a unique Cas9:sgRNA electroporation experiment. Mean ± SD, with Student’s two-sided t-test.***P < 0.001, ****P < 0.0001. G–J Human CD34 + HSPCs from three healthy donors were edited by Cas9 RNP electroporation (EP) targeting a neutral locus and S1PR3 coding sequence infused into NBSGW mice 24 h after electroporation. After 12 weeks, engrafted bone marrow was characterized by immunophenotyping. G Indels determined by Sanger sequencing before transplantation. (H––J) Quantification of different human cell types between the neutral locus and S1PR3 targeting group. Human chimerism, hCD45 + ; Monocytes, hCD45 + CD33 + SSClowCD14 + ; neutrophil, hCD45 + CD33 + SSChighCD16 + . N = 3 independent biological replicates, each replicate indicates one mouse. Mean ± SD, 2-sided Mann-Whitney test. *P = 0.025, **P = 0.004.