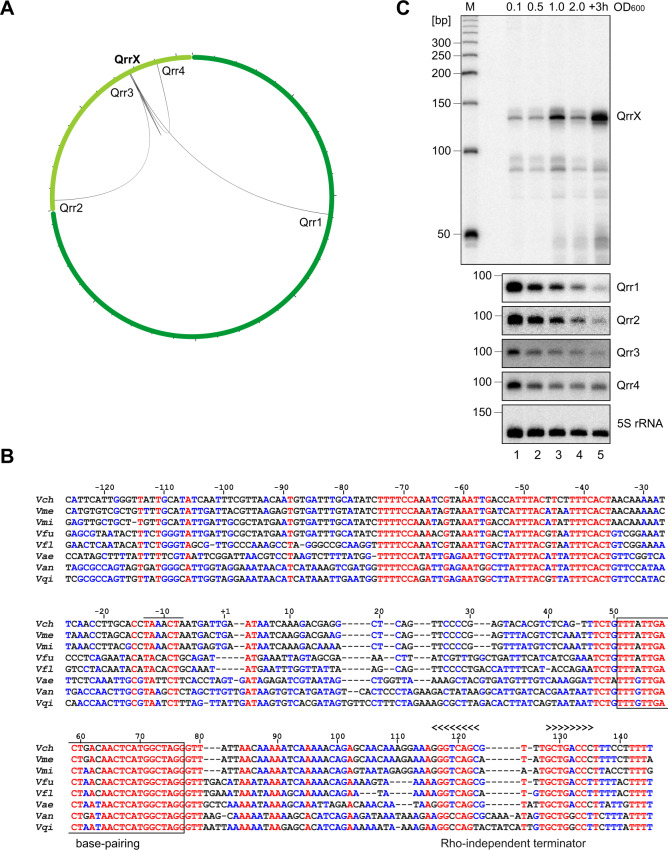
Fig. 2. Identification of QrrX sponge RNA.
A Interaction partners of QrrX. Circos plot visualizing interaction partners of QrrX identified by RIL-seq. The first and the second chromosome are marked in dark and light green, respectively. Circos plot was generated using the circos component of the Dash Bio package. B Alignment of qrrX sequences from various Vibrio species. The qrrX sequences including the promotor regions were aligned using the Multalin tool65. The predicted base-pairing region and the Rho-independent terminator are indicated. Numbers above the sequences indicate the distance to the transcriptional start site. Vch, Vibrio cholerae; Vme, Vibrio metoecus, Vmi, Vibrio mimicus; Vfu, Vibrio furnissii; Vfl, Vibrio fluvialis; Vae, Vibrio aestuarianus; Van, Vibrio anguillarum, Vqi, Vibrio qinghaiensis. C Expression of QrrX. V. cholerae wild-type cells were cultivated in LB medium and RNA samples were collected at various stages of growth. Northern blot analysis using specific oligonucleotide probes was performed to determine QrrX, Qrr1, Qrr2, Qrr3, and Qrr4 levels. Probing for 5S ribosomal RNA served as loading control. The experiment was done in three independent biological replicates. Source data underlying panel C are provided as a Source Data file.