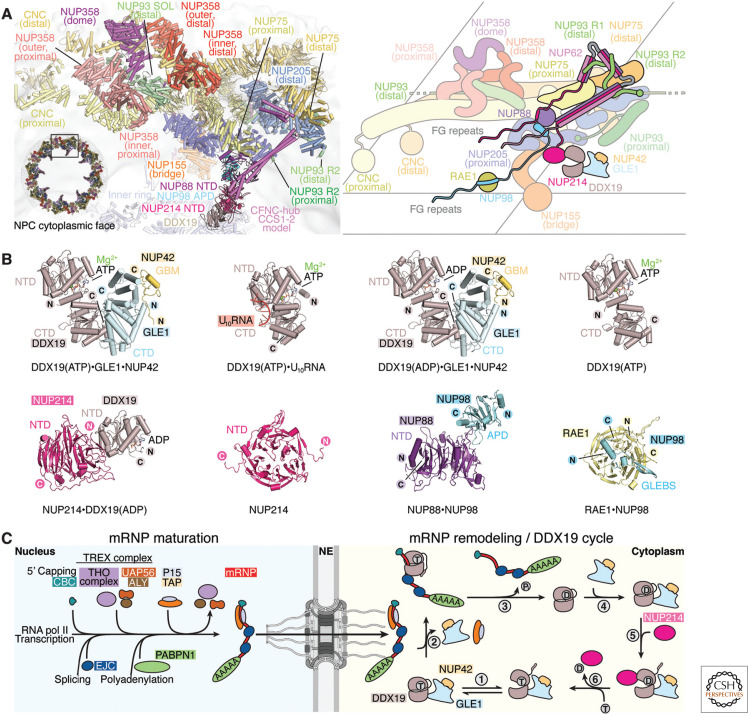
Figure 4.
mRNP export and remodeling at the cytoplasmic face of the nuclear pore complex (NPC). (A) Cartoon representation and corresponding schematic conceptualization of a spoke of the NPC cytoplasmic face illustrating the relative positions of the cytoplasmic filament nucleoporins, with the cytoplasmic filament nucleoporin complex (CFNC) anchored by NUP93R1 and a pentameric bundle of NUP358 bound to the stalk of tandem-arranged Y-shaped coat nucleoporin complexes (CNCs). (B) Cartoon representations of crystal structures of human cytoplasmic filament complexes: DDX19(ADP)•GLE1CTD•NUP42GBM (PDB ID 6B4I) (Lin et al. 2018), DDX19(ATP)• GLE1CTD•NUP42GBM (PDB ID 6B4J) (Lin et al. 2018), DDX19(ATP)•U10RNA (PDB ID 3FHT) (von Moeller et al. 2009), DDX19(ATP) (PDB ID 6B4K) (Lin et al. 2018), DDX19(ADP)•NUP214NTD (PDB ID 3FMO) (Napetschnig et al. 2009), NUP214NTD (PDB ID 2OIT) (Napetschnig et al. 2007), NUP88NTD•NUP98APD (PDB ID 7MNI) (Bley et al. 2022), RAE1•NUP98GLEBS (PDB ID 3MMY) (Ren et al. 2010). (C) Schematic representation of the maturation, export, and remodeling of mRNPs by the DDX19 helicase cycle. In the nucleus, mRNA is transcribed by RNA polymerase II, followed by 5′ capping, recruitment of the cap-binding complex (CBC), deposition of exon junction complexes (EJCs) at splice sites, loading of the transcription-export (TREX) complex in a splicing-dependent manner, 3′ polyadenylation, and deposition of polyadenylate-binding nuclear protein 1 (PABPN1). ATP-hydrolyzing DEAD-box helicase UAP56 may facilitate loading of export factors P15•TAP to produce an export-competent mRNP. The exported mRNP is remodeled at the cytoplasmic face of the NPC, where P15•TAP is removed in a DDX19-dependent manner: (1) ATP-bound DDX19 cycles between autoinhibited and closed conformations. (2) Upon binding RNA, DDX19 adopts a closed, catalytically active conformation that is incompatible with GLE1 binding but could strip P15•TAP from mRNA. (3) ATP hydrolysis by DDX19 triggers RNA release, converting DDX19 to an autoinhibited ADP-bound conformation in which the autoinhibitory α-helix binds between amino- and carboxy-terminal RecA domains. (4) GLE1 binding destabilizes the autoinhibited conformation. (5) NUP214 binding converts DDX19 to an open conformation, promoting nucleotide exchange. (6) Nucleotide exchange displaces NUP214, priming the DDX19(ATP)•GLE1•NUP42 complex to restart the cycle (Lin et al. 2018). (Figure adapted from Bley et al. 2022, with permission from the authors.)