In this study, Nie et al. performed proteome profiling with label-free quantification in control and single/double-KO cells of USP7 and its closest homolog, USP47, and for the first time revealed the proteome changes caused by USP7 and/or USP47 depletion. Their proteome-wide analysis uncovered potential USP7 substrates, which provides a resource for further functional studies.
Keywords: USP7, proteomics, ubiquitination, MGA
Abstract
Deubiquitylating enzymes (DUBs) remove ubiquitin chains from proteins and regulate protein stability and function. USP7 is one of the most extensively studied DUBs, since USP7 has several well-known substrates important for cancer progression, such as MDM2, N-MYC, and PTEN. Thus, USP7 is a promising drug target. However, systematic identification of USP7 substrates has not yet been performed. In this study, we carried out proteome profiling with label-free quantification in control and single/double-KO cells of USP7and its closest homolog, USP47. Our proteome profiling for the first time revealed the proteome changes caused by USP7 and/or USP47 depletion. Combining protein profiling, transcriptome analysis, and tandem affinity purification of USP7-associated proteins, we compiled a list of 20 high-confidence USP7 substrates that includes known and novel USP7 substrates. We experimentally validated MGA and PHIP as new substrates of USP7. We further showed that MGA deletion reduced cell proliferation, similar to what was observed in cells with USP7 deletion. In conclusion, our proteome-wide analysis uncovered potential USP7 substrates, providing a resource for further functional studies.
Protein ubiquitination is an important post-translational modification, which plays a prominent role in a variety of cellular pathways (Ciechanover 1998). Dysregulation of the ubiquitination process has been implicated in various human diseases, including cancer, infection, and neurodegeneration (Tan et al. 2007; Isaacson and Ploegh 2009; Senft et al. 2018). Ubiquitination majorly controls protein degradation to maintain the homeostasis of the proteome (Hershko and Ciechanover 1998). Protein ubiquitination and deubiquitylation are controlled by two sets of enzymes. While the E3 ubiquitin ligases are the key enzymes responsible for the conjugation of ubiquitin to protein substrates, deubiquitylating enzymes (DUBs) are proteases that cleave ubiquitin chains from proteins (Sun 2008). DUBs play critical roles in protein degradation and ubiquitin recycling (D'Arcy et al. 2015). Approximately 100 DUBs encoded by the human genome can be grouped into seven subclasses that are structurally distinct (Lange et al. 2022). Previously, five subclasses of DUBs were classified, with ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), and Machado–Joseph disease proteases (MJDs) as cysteine-based DUBs, and JAB1/MPN/Mov34 (JAMMs) as zinc-binding metalloproteases (Lange et al. 2022). Recently, two new DUB subclasses, MINDY and ZUP DUBs, were defined as cysteine-based DUBs, which are highly selective at cleaving K48- and K63-linked polyubiquitin chains, respectively (Abdul Rehman et al. 2016; Kwasna et al. 2018). Among these DUBs, USPs are the largest subclass. Ubiquitin-specific peptidase 7 (USP7), also known as herpes-associated ubiquitin-specific protease (HAUSP), belongs to this subclass. USP7 is one of the most extensively studied DUBs, as many proteins involved in cell cycle, DNA repair, chromatin remodeling, and epigenetic regulation are identified as its substrates (Bhattacharya et al. 2018; Peng et al. 2019).
USP7 was originally identified as an interaction protein of herpes simplex virus type 1 (HSV1) ICP0, and later as an interaction protein of other viral proteins such as Epstein–Barr virus (EBV) EBNA1 (Everett et al. 1997; Holowaty et al. 2003b; Daubeuf et al. 2009). Since then, more and more proteins have been identified as interaction proteins of USP7, which suggests that USP7 may be involved in multiple cellular processes, such as cell cycle, DNA repair, chromatin remodeling, and epigenetic regulation. Aberrant gain or loss of function of USP7 may be related to several human diseases. For example, USP7 was found to be overexpressed in multiple cancers, and its overexpression contributes to tumor progression (Song et al. 2008; Cheng et al. 2013; Wang et al. 2016; Carrà et al. 2017). Additionally, children with USP7 mutations or deletions suffer from neurodevelopmental disorders (Fountain et al. 2019; https://www.usp7.org). Although USP7 was first identified in the nucleus, later studies suggest that USP7 has a dynamic overall cellular distribution, which may depend on pathological conditions and genetic perturbations (Everett et al. 1997; Becker et al. 2008).
USP7 protects substrates from degradation by removing ubiquitin chains on its substrates. USP7 contains an N-terminal MATH domain, a central catalytic domain (CD), and five C-terminal tandem ubiquitin-like (Ubl) domains (Holowaty et al. 2003a; Faesen et al. 2011; Pozhidaeva and Bezsonova 2019). Studies show that different substrates may bind to different USP7 domains; for example, MDM2 and TP53 bind to the N-terminal domain of USP7, while DNMT1 and UHRF1 interact with its C-terminal Ubl domains (Sheng et al. 2006; Cheng et al. 2015; Zhang et al. 2015). USP7 may regulate many critical cellular processes via the large number of identified USP7 substrates, such as TRIM27 in immune response, DAXX and PTEN in tumor suppression, DNMT1 in epigenetics, and HLTF and UVSSA in DNA damage (Wang et al. 2019). Of these substrates, arguably the most prominent one is MDM2, the E3 ligase for TP53, which supports the development of USP7 inhibitors, including FT671, XL177A, and GNE-6640 (Kategaya et al. 2017; Turnbull et al. 2017; Schauer et al. 2020). However, TP53 is also a substrate of USP7 (Li et al. 2002), which may make the connection between USP7 and MDM2/TP53 complicated. Moreover, Tp53 deletion cannot rescue embryonic lethality in Usp7 knockout mice (Kon et al. 2010; Agathanggelou et al. 2017). In order to further study the functions of USP7 and support the development of USP7-specific inhibitors, it is critical to reveal the key and the wide-ranging substrates of USP7.
USP47 is another DUB and the closest homolog of USP7, which shares similar domain structures with and has 48.4% similarity to USP7 in their catalytic domains (Catic et al. 2007; Palazón-Riquelme et al. 2018). USP47 may also be an oncology target, since it deubiquitinates DNA polymerase β (POLB) to regulate DNA base excision repair (BER) (Parsons et al. 2011). USP7 and USP47 are known to work together to deubiquitinate NLRP3, which is required for inflammasome activation, although USP7 and USP47 show different enzymatic properties (Piao et al. 2015; Palazón-Riquelme et al. 2018). Furthermore, some earlier USP7 inhibitors (e.g., P22077 and P005091) inhibit both USP7 and USP47 (Tian et al. 2011; Weinstock et al. 2012). However, until now, the substrates of USP47 remained limited. It will be interesting to uncover the substrates of USP7 and USP47 and determine whether they work together on a subset of these substrates.
To reveal globally the substrates of USP7, we performed quantitative proteomics analysis using control, USP7 KO, USP47 KO, and USP7/USP47 DKO cells. These analyses, together with transcriptome and protein–protein interaction studies, revealed 20 high-confidence substrates of USP7, including two new substrates—MGA and PHIP—that we experimentally validated in this study. Additionally, we showed that deletion of USP7 or USP47 led to distinct changes in proteomes, arguing that these two DUBs have different functions. Furthermore, we showed that although current USP7 inhibitors FT671 and GNE-6640 were able to regulate USP7 substrates, they exerted similar cytotoxicity in several paired wild-type and USP7 KO cell lines, implying that they may have other targets besides USP7.
Results
The proteome of USP7 KO cells diverges from that of USP47 KO cells
Given its higher expression in a variety of cancers (Supplemental Fig. S1A), USP7 is becoming one of the most extensively studied deubiquitinates (Lu et al. 2021). USP7 has a distinct domain structure among USPs, which includes an N-terminal MATH domain, a central catalytic domain (CD), and five C-terminal tandem ubiquitin-like (Ubl) domains (Supplemental Fig. S1B; Rougé et al. 2016). USP47 is the closest homolog to and shares similar structure with USP7 (Supplemental Fig. S1B; Palazón-Riquelme et al. 2018). To understand comprehensively the proteomes regulated by USP7 and USP47, HEK293A-derived USP7 KO, USP47 KO, and USP7/USP47 DKO cells were generated by CRISPR/Cas9 genome-editing technology (Fig. 1A; Supplemental Fig. S2). Moreover, we confirmed the change of known USP7 substrates, such as MDM2, in USP7 KO cells (Supplemental Fig. S2B). USP7 KO cells showed defective cell proliferation (Fig. 1B), which is consistent with previous results (Shan et al. 2018; Jin et al. 2019; Peng et al. 2019). Of note, USP47 KO did not affect cell proliferation, while USP7/USP47 DKO led to reduced cell proliferation (Supplemental Fig. S3A,B). Furthermore, USP7 KO also affected cell proliferation in A549 and HeLa cells (Supplemental Fig. S3C).
Figure 1.
Label-free quantitative proteomics analysis of USP7 and USP47 single- and double-knockout cells. (A) Whole-cell lysates prepared from control HEK293A, USP7 KO, USP47 KO, and USP7/USP47 DKO were immunoblotted with the indicated antibodies. (B) The proliferation curve of parental and USP7 KO cells. Experiments with each cell line were repeated three times. (C) The workflow of the label-free quantitative proteomics analysis. (D) The number of total quantified proteins of each replicate. (E) The fold change of reported USP7 substrates in USP7 and USP47 KO cells when compared with those in 293A cells. The list of reported USP7 substrates was generated based on a previous study (Pozhidaeva and Bezsonova 2019).
We conducted an MS-based label-free quantitative proteomics analysis to profile the proteomes in each of these KO cells (Fig. 1C). We performed four biological replicates in each of the control and KO cells; a total of 10,300 protein groups and an average of 8000 proteins in each replicate were identified, and >6000 proteins were consistently identified in all conditions (Fig. 1D; Supplemental Table S1). As expected, several known substrates of USP7 were reduced in USP7 KO and USP7/USP47 DKO cells but not in USP47 KO cells (Fig. 1E; Supplemental Table S2; Pozhidaeva and Bezsonova 2019).
USP47 not only shares a similar catalytic domain with USP7, but also has similar C-terminal multiple Ubl domains (Supplemental Fig. S1B). We speculated that USP7 and USP47 may have overlapping substrates. However, principal component analysis clearly revealed that these samples could be separated into two groups: 293A and USP47 KO, USP7 KO, and USP7/USP47 DKO (Fig. 2A). This implies that USP7 deletion was the main driver to separate these samples, with limited contribution from USP47 KO. Furthermore, unsupervised clustering showed patterns similar to those of principal component analysis, and these samples showed high reproducibility (Pearson's r > 0.95) (Fig. 2B). As mentioned previously, several known substrates were reduced/destabilized in USP7 KO (Fig. 1E). Furthermore, we examined the fold change of proteins in each KO samples. In line with the above results, results from USP7 KO correlated highly with those from USP7/USP47 DKO (Fig. 2C), while USP47 KO showed restricted correlation with the other two (Supplemental Fig. S4A,B). Collectively, these data not only demonstrate the high quality of our proteome-profiling results, but also indicate that USP7 differs from USP47 and that USP47 has very few substrates in the cell.
Figure 2.
USP7 KO and USP47 KO cells show different proteome characteristics. (A) The two-dimensional principle component analyses separated samples into two parts; i.e., wild type and USP47 KO versus USP7 KO and USP7/USP47 DKO. (B) Unsupervised clustering of results obtained in USP7 KO and USP7/USP47 DKO cells, based on Pearson correlation among biological replicates. (C) The correlation of fold change in USP7 KO and USP7/USP47 DKO versus control 293A cells. Each dot denotes a protein. The orange dots indicate reported USP7 substrates identified in this study.
Identification of candidate USP7 substrates
To uncover potential substrates of USP7, proteins with two or more unique peptides were subjected to a Student's t-test with a permutation-based false discovery rate of <0.05 and S0 = 0.1 using Perseus software (Tyanova and Cox 2018). In USP7 KO cells, 144 and 320 proteins were significantly decreased or increased, respectively (Fig. 3A; Supplemental Table S3). Unexpectedly, the level of TP53 did not change (see Supplemental Fig S2B), although we identified some known substrates of USP7—such as TRIM27, DHX40, and DAXX—that have reduced expression in USP7 KO cells. Gene set enrichment analysis (GSEA) showed that some proteins involved in metabolism were significantly reduced, including components of the ribosome, oxidative phosphorylation, and protein export, while proteins involved in arginine and proline metabolism and the PPAR signaling pathway were significantly up-regulated (Fig. 3B). However, we could not exclude the possibility that these altered metabolism pathways may be indirect changes due to reduced cell proliferation. Besides the known USP7 substrates and their associated complexes, such as TRIM27, GMPS, and DAXX, the polycomb-repressive complex (PRC), respiratory chain complex, and interferon complex that were down-regulated were also revealed by the protein–protein interaction networks using the STRING database (Fig. 3C). Further molecular function enrichment analysis of these changed proteins agrees with the results revealed by protein–protein interaction networks (Fig. 3C). These analyses indicate that these cellular processes are significantly altered in USP7 KO cells.
Figure 3.
Analysis of proteome changes in USP7 KO versus control cells. (A) Volcano plot showing the differentially expressed proteins in USP7 KO versus parental 293A cells. Each dot denotes a protein. The brown and olive dots represent significantly up-regulated and down-regulated proteins, respectively. (B) Gene set enrichment analysis (GSEA) was conducted with a MSigDB KEGG subset based on the preranked protein list of USP7 KO versus 293A cells. (C) The significantly down-regulated proteins in USP7 KO cells were subjected to protein–protein interaction (PPI) analysis with STRING. The top panel shows the PPI network. The bottom panel shows the significant functional enrichment of the significantly down-regulated proteins. (D) The correlation between the protein changes and mRNA changes of each gene product in USP7 KO versus 293A.
USP7 is known to stabilize several transcription factors, such as TP53, FOXOs, and PTEN, implying the involvement of USP7 in transcription regulation (Pozhidaeva and Bezsonova 2019). Our molecular function enrichment also indicated the role of USP7 in transcription regulation (Fig. 3C). To eliminate any proteins that may be regulated indirectly by USP7 via its role in transcription regulation, we compared transcriptomes and proteomes in control and USP7 KO cells (Fig. 3D). Unexpectedly, several top decreased proteins, such as MAP7D2, IFIT3, and ISG15, were also changed at their mRNA levels (Fig. 3D). These data indicate that some of these proteins that are decreased in USP7 KO cells may be reduced via transcriptional regulation and therefore are not direct USP7 substrates.
As mentioned above, USP7 KO and USP7/USP47 DKO showed similar proteomes. We conducted the same analyses to reveal the proteins changed in USP7/USP47 DKO cells. In USP7/USP47 DKO cells, 643 and 517 proteins were significantly decreased or increased, respectively (Supplemental Fig. S5A; Supplemental Table S4). Again, the level of TP53 did not change in USP7/USP47 DKO cells, which is the same as what was observed in USP7 KO cells (see Supplemental Fig S2B). GSEA showed similar enriched pathways in USP7/USP47 DKO cells when compared with those in USP7 KO cells (Fig. 3B; Supplemental Fig. S5B). Similarly, the protein–protein interaction networks of down-regulated proteins also revealed the PRC complex and respiratory chain complex (Supplemental Fig. S5C), suggesting that these are the common complexes and that cellular processes changed in both USP7 KO and USP7/USP47 DKO cells.
Validation of changed proteins by USP7 reconstitution in USP7 KO cells
To further validate protein changes observed in USP7 KO cells, we confirmed their alternations by Western blot analysis (Fig. 4A; Supplemental Fig. S6A). CKMT1A is the top increased protein at both the transcription and protein levels, which were used as a positive control in these experiments. Reconstitution of USP7 in USP7 KO cells reversed protein alternations, including those affected at mRNA levels, such as ISG15 and IFIT3. To further confirm that these protein changes are not specific for 293A cells, we also validated some of these proteins in A549 and HeLa cells (Fig. 4B; Supplemental Fig. S6B). Similarly, reconstitution of USP7 rescued the level of proteins such as MGA and PHIP in both A549 and HeLa USP7 KO cells (Supplemental Fig. S6C). Furthermore, USP7 overexpression caused accumulation of proteins such as MGA, PHIP, and the well-known USP7 substrate MDM2 (Fig. 4C). Moreover, FT671 is a selective USP7 inhibitor developed based on cocrystal structures (Turnbull et al. 2017). FT671 can destabilize MDM2 in HCT116 cells. We confirmed that FT671 and GNE-6640 treatment reduced the expression of MDM2, a well-known substrate of USP7, in a dose-dependent manner (Fig. 4D; Supplemental Fig. S6D). It also reduced the expression of other potential USP7 substrates such as MGA and PHIP (Fig. 4D; Supplemental Fig. S6D), although FT671 appeared to be more efficient at reducing MGA than GNE-6640. Altogether, these data indicate that MGA and PHIP may be generally regulated by USP7, regardless of cellular background.
Figure 4.

Reconstitution of USP7 rescues protein changes and proliferation defects in USP7 KO cells. (A) Immunoblots of whole-cell extracts prepared from 293A, USP7 KO, and USP7 KO cells stably expressing USP7 with the indicated antibodies. Vinculin was used as a loading control, while CKMT1A was also included as a control. (B) The candidate proteins from A were further validated in A549 WT, A549 USP7 KO, HeLa WT, and HeLa USP7 KO cells. (C) Immunoblotting was conducted with the indicated antibodies in extracts prepared from 293A, A549, HeLa, and corresponding cells with USP7 overexpression. (D) Immunoblot of PHIP, MGA, CUL4A, and L3MBTL2 in 293A cells treated with the indicated concentrations of FT671 for 48 h. Actin was used as a loading control, and MDM2 was used as a positive control. (E) The proliferation curve of 293A, USP7 KO, and USP7 cells stably expressing wild-type USP7 or a catalytically inactive mutant of USP7.
As mentioned above, USP7 KO cells showed defective cell proliferation (Fig. 1B). As controls, we showed that reconstitution with wild-type USP7, but not a USP7 catalytically inactive mutant (USP7C223S), rescued cell proliferation defects (Fig. 4E). Also, this USP7 catalytically inactive mutant of USP7 could not rescue the decreased protein levels of several potential USP7 substrates, such as MGA and PHIP (Supplemental Fig. S6E)
MGA and PHIP are novel substrates of USP7
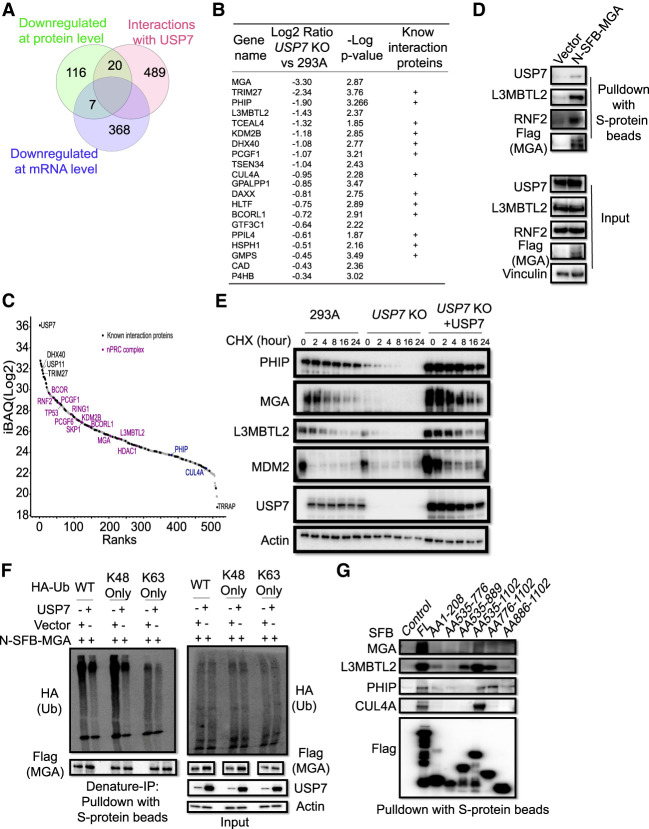
To further define the substrates of USP7, we overexpressed N-terminal SFB-tagged USP7 and then used our tandem affinity purification strategy to identify potential USP7 interaction proteins (Supplemental Fig. S7A; Supplemental Table S5; Srivastava et al. 2018). We reasoned that USP7 substrates should change at the protein level but not at the mRNA level. Moreover, although USP7 may only associate with its substrates transiently, the interaction with USP7 still increases the chance of these proteins being direct substrates of USP7. Therefore, we compiled lists of proteins down-regulated at the protein level and/or at the mRNA level in USP7 KO cells and compared them with the proteins identified as USP7-interacting proteins (Fig. 5A). A total of 20 high-confidence substrates of USP7 was identified (Fig. 5A,B). Some known USP7 substrates and interaction proteins are included, such as TRIM27, DHX40, and GMPS (Fig. 5B), which were previously identified as strong interactors and substrates of USP7 (Georges et al. 2018, 2019). Two top candidate USP7 substrates are MGA and PHIP (Fig. 5B).
Figure 5.
MGA is a novel high-confidence substrate of USP7. (A) Venn diagram for down-regulated proteins, down-regulated mRNAs, and high-confidence USP7 interaction proteins. (B) The proteins from the overlapping down-regulated proteins in USP7 KO cells and those identified as USP7-interacting proteins. These proteins are ranked by the log2 ratio of their abundance in USP7 KO versus 293A cells. (C) Potential USP7-interacting proteins were ranked by intensity-based absolute quantification (iBAQ), identified in this study. Known interaction proteins annotated in the CRAPome are labeled in black. The DHX40 and TRIM27 were used as positive controls. (D) Coimmunoprecipitation of USP7 with MGA. MGA was fused with N-terminal S-protein, FlAG, and streptavidin-binding peptide (SFB) tags. L3MBTL2 and RNF2 were used as positive controls. (E) Immunoblotting assay of PHIP, MGA, and L3MBTL2 stability in 293A, USP7 KO, and USP7 KO cells stably expressing WT USP7. Cells were treated with 50 µg/mL cycloheximide for the indicated times. (F) USP7 deubiquitinates wild-type ubiquitin and “Lys-48”-linked ubiquitin chains from MGA. HEK293T was transfected with vector or the indicated plasmid together with constructs encoding WT or lysine-specific mutants (K48 only or K63 only) of HA-ubiquitin. MGA was recovered with S-protein beads under denaturing conditions. (G) HEK293T cells were transfected with constructs encoding full-length SFB-tagged USP7 or its deletion variants and then subjected to coprecipitation with S-protein beads. Endogenous MGA, PHIP, L3MBTL2, and CUL4A coimmunoprecipitated with SFB-tagged USP7 or its deletion variants were detected by immunoblotting with the indicated antibodies. (FL) Full-length, (AA) amino acid.
MGA, a MAX gene-associated protein, was one of the top decreased proteins in USP7 KO cells in our proteome profiling (Fig. 3A). MGA is the largest member of the MAX-interacting transcription factor network and functions as a dual-specificity transcription factor with two different DNA binding domains: a T domain at the N terminus and a basic helix–loop–helix zipper (bHLHZip) domain at the C terminus (Hurlin et al. 1999). MGA may function as a tumor suppressor, as MGA loss-of-function mutations were identified in lung cancer and leukemia (De Paoli et al. 2013; The Cancer Genome Atlas Research Network 2014). MGA, together with L3MBTL2, PCGF6, E3F6, TFDP1, and others, comprises the noncanonical PRC1.6 complex (Stielow et al. 2018). MGA may act as a scaffold for PRC1.6 assembly and guide PRC1.6 to specific genomic targets (Qin et al. 2021). USP7 was reported previously to interact with some PRC subunits (Maat et al. 2021; Su et al. 2021). Moreover, several members of this PRC1.6 complex were uncovered in our USP7 interactome and/or list of down-regulated proteins, such as L3MBTL2 and PCFG6 (Fig. 5B,C). PHIP, the WD-40-containing protein, is a substrate receptor of the CUL4A/B E3 ligase complex (Morgan et al. 2017; Jang et al. 2018). Furthermore, CUL4A and DDB1, members of the CUL4A/B E3 ligase complex, were down-regulated in USP7 KO cells and were shown to interact with USP7 (Figs. 3C, 5C). XL177A, a highly potent and selectively irreversible USP7 inhibitor, also significantly decreased the level of MGA and PHIP (Schauer et al. 2020; Bushman et al. 2021). Additionally, another data-independent acquisition (DIA)-MS study showed that two other USP7 inhibitors (FT671 and GNE-6640) not only decreased MGA and PHIP protein levels, but also increased the ubiquitination level of MGA and PHIP (Steger et al. 2021). Collectively, these data suggest that MGA and PHIP are likely candidate substrates of USP7.
We overexpressed N-terminal SFB-tagged MGA or PHIP and showed that they could pull down endogenous USP7 and other proteins in their respective complexes, such as L3MBTL2 and CUL4A (Fig. 5D; Supplemental Fig. S7B). As shown in Figure 5E, treatment with the translation inhibitor cycloheximide led to a rapid decline of MGA and PHIP in USP7 KO cells but not in wild-type or USP7 reconstitution cells. These proteins showed patterns similar to that of MDM2, which is a well-known substrate of USP7. To further determine whether USP7 directly affects the ubiquitination status of MGA and PHIP, cells expressing MGA or PHIP were transfected with constructs encoding WT or lysine mutants (K48 only or K63 only) of HA-ubiquitin, together with USP7 or vector. MGA or PHIP was pulled down by S-protein beads under denaturing conditions. USP7 expression reduced wild-type, Lys-48-linked, and even Lys-63-linked ubiquitination of both MGA and PHIP (Fig. 5F; Supplemental Fig. S7C). Additionally, MG132 treatment partially restored the protein levels of MDM2, MGA, PHIP, and L3MBTL2 (Supplemental Fig. S7D). Taken together, these data indicate that the protein levels of MGA and PHIP regulated by USP7 are dependent on the ubiquitination-mediated degradation pathway.
USP7 harbors an N-terminal MATH domain, a central catalytic domain (CD), and five C-terminal tandem ubiquitin-like (Ubl) domains (Supplemental Fig. S1B; Rougé et al. 2016). Different substrates may bind to different USP7 domains (Kim and Sixma 2017). We constructed different N-terminal-tagged USP7 truncation mutants (Supplemental Fig. S7E; Pfoh et al. 2015) and used these mutants to pull down endogenous proteins. These results showed that endogenous MGA, PHIP, and their associated proteins could interact with full-length USP7, and these interactions are mainly mediated by the Ubl domains of USP7 (Fig. 5G). Together, these data indicate that MGA and PHIP are likely USP7 substrates.
MGA KO led to cell proliferation defects
As mentioned above, we validated MGA as a novel substrate of USP7. MGA is a poorly understood transcription factor. To further study MGA as a substrate of USP7, we generated MGA KO and MGA/USP7 DKO cells, which were validated by Western blot and sequencing (Supplemental Fig. S8A,B). MGA is indispensable for pluripotent ICM cells and the growth of ESCs (Washkowitz et al. 2015). A previous study showed that MGA deletion reduced cell proliferation (Stielow et al. 2018). Consistent with the previous report, MGA KO showed reduced cell proliferation (Fig. 6A). However, USP7 KO and USP7/MGA DKO showed more severe reduction in cell proliferation when compared with MGA KO cells (Fig. 6A).
Figure 6.
MGA KO cells only show mild growth defects when compared with USP7 KO cells. (A) Proliferation of 293A, USP7 KO, MGA KO, and MGA/USP7 DKO cells was measured. Experiments for each cell line were repeated three times. (B) The two-dimensional principle component analyses separated samples into two parts (i.e., WT and MGA KO vs. USP7 KO and USP7/MGA DKO) based on RNA-seq data. (C) Unsupervised clustering of different genes/proteins in three knockout cell lines based on RNA-seq data. (D) Volcano plot showing the differentially expressed mRNAs in USP7 KO versus 293A cells. Each dot denotes an mRNA. The brown and olive dots represent significantly up-regulated and down-regulated genes/proteins, respectively. (E) GSEAs of transcripts in USP7 KO versus 293A cells were conducted against MSigDB KEGG and hallmark subsets.
To further characterize the difference between MGA KO and USP7 KO cells, we compared the transcriptomes in these cells (Supplemental Table S6). Unexpectedly, control wild-type cells and MGA KO cells were clustered together (Fig. 6B), suggesting that MGA KO only led to minor changes in gene transcription. On the other hand, UPS7 KO and USP7/MGA DKO were clustered together and showed similar transcription/mRNA patterns (Fig. 6C). Furthermore, we examined the fold change of genes in these RNA-seq data. In line with the above-mentioned results, data from USP7 KO correlated highly with those from USP7/MGA DKO, while MGA KO showed restricted correlation with USP7 KO (Supplemental Fig. S8C). Thus, the functions of USP7 are likely mediated by many of its substrates, and MGA is only one of these substrates. To further support this hypothesis, we knocked down PHIP by shRNA to a level similar to that observed in USP7 KO cells (Supplemental Fig. S9A). PHIP down-regulation also reduced cell proliferation; however, USP7 KO cells still displayed more severe defects in cell proliferation (Supplemental Fig. S9B). Similar results were also observed in A549 cells (Supplemental Fig. S9C,D), indicating that both MGA and PHIP contribute to cell proliferation. Altogether, these results suggest that the functions of USP7, at least in cell proliferation, are mediated by many of its substrates.
Immune response is potentially inhibited by USP7 KO
Increasing evidence suggests that USP7 may regulate immune response by stabilizing a number of USP7 substrates. However, there are contradictory data as to precisely how USP7 may influence the immune response. For example, a previous report suggests that inhibition of USP7 leads to ubiquitination of NF-κB, which down-regulates TLR- and TNFR-induced expression of interleukins (IL-6) and TNFα (Colleran et al. 2013). USP7 also positively regulates Treg cell-induced suppression of autoimmune responses in vitro and in vivo (van Loosdregt et al. 2013), and chemical inhibition of USP7 leads to abrogated inflammasome formation (Palazón-Riquelme et al. 2018). On the other hand, other data suggest that USP7 and TRIM27 negatively regulate antiviral type I interferon signaling (Cai et al. 2018).
To gain further insights into the potential immune response regulated by USP7, we analyzed the transcription changes in USP7 KO cells (Fig. 6D). Interestingly, the interferon regulation genes were the top decreased genes in USP7 KO cells, which include IFIT2, IFIT3, ISG15, IRF5, and IRF6 (Fig. 6D). Moreover, IFIT3 and ISG15 were also the top decreased proteins in our proteome profiling (Fig. 3A). Furthermore, GSEA with KEGG and hallmark pathway analysis showed that immune response pathways were decreased in USP7 KO cells (Fig. 6E). To investigate potential transcription factors (TFs) responsible for the down-regulation of interferon regulation genes, we used ChEA3 to analyze the decreased gene transcription due to USP7 loss (Supplemental Fig. S9E). As expected, the top enriched TFs, such as IRF7, SP140, and BATF2, were those responsible for the regulation of interferon genes, which cover almost all of the decreased interferon regulation genes observed in our data set (Supplemental Fig. S9E). Similar results were reported (Colleran et al. 2013; Liu et al. 2018), although we and others have not yet been able to establish any direct or indirect relationship between USP7 and TFs such as IRF7. Additionally, an early study indicated that USP7 may be a positive or negative regulator of these immune-related genes, depending on subcellular localization and/or contexts (Liu et al. 2018). However, our results suggest that USP7 plays a positive role in regulating immune response.
Paired control and USP7 KO cells revealed potential context-dependent effects of USP7 inhibitors
Given that USP7 may be involved in multiple cellular processes, there is a lot of interest in developing USP7 inhibitors as potential therapeutic agents. Thanks to the cocrystal structures of USP7, a number of new specific USP7 inhibitors were developed recently, which include FT671, GNE-6640, and XL177A (Kategaya et al. 2017; Turnbull et al. 2017; Schauer et al. 2020). The decreased MDM2 and increased TP53 were used as biomarkers to test these inhibitors. However, Tp53 deletion cannot rescue embryonic lethality in Usp7 knockout mice (Kon et al. 2010; Agathanggelou et al. 2017). Thus, USP7 may play critical roles independent of TP53. In this study, we used three cell lines. While HEK293A and A549 may express wild-type TP53, TP53 function is impaired in HeLa cells due to the targeted degradation of TP53 by HPV E6 in these cells.
In theory, the loss of target will counteract the cytotoxicity of target inhibitors. Thus, we treated several of our paired wild-type and USP7 KO cells with FT671 or GNE-6640. Unexpectedly, we showed that the cytotoxicity of these inhibitors showed no or minor difference in parental or USP7 KO cells in HEK293A, A549, and HeLa cell lines, regardless of whether these cells have functional TP53 (Fig. 7). Furthermore, to avoid any potential adaptation due to continuous USP7 depletion in these cells, we transiently knocked down USP7 by shRNA in 293A and A549 and showed that the cytotoxicity of these inhibitors remained the same or similar in parental and these knockdown cells (Supplemental Fig. S10). Thus, these two USP7 inhibitors did not display a robust antiproliferation effect in these three paired cell lines, implying that these USP7 inhibitors may only be effective in specific genetic and/or tissue contexts.
Figure 7.
USP7 inhibitors in parental and USP7 KO cells. (A,B) CellTiter-Glo assay revealed that USP7 KO cells showed similar sensitivity to USP7 inhibitors FT671 (A) and GNE-6440 (B) when compared with that in control parental cells. Cells were exposed to the indicated concentrations of inhibitors and cultured for 3 d. Data are from three technical replicates.
Discussion
In this study, we generated USP7 KO, USP47 KO, and USP7/USP47 DKO cells and revealed different proteomes in USP7 KO and USP47 KO cells via in-depth label-free quantitative proteomics analysis. Because of the mild changes of proteomes in USP47 KO cells, we focused our attention on USP7 KO cells. Unexpectedly, the polycomb-repressive complex (PRCs), respiratory chain complex, and interferon complex are among the top down-regulated proteins in USP7 KO cells. To further identify USP7 substrates, we compared protein-profiling data with RNA-seq data. Interestingly, some of the top down-regulated proteins identified by proteome profiling, such as MAP7D2, IFIT3, and ISG15, are also the top down-regulated genes at their mRNA levels, suggesting that USP7 likely controls proteomes in part via its involvement in transcriptional regulation. Indeed, our further bioinformatics analysis implied that USP7 may be required for proper immune response in the cell.
In order to obtain the high-confidence substrates of USP7, we also performed tandem affinity purification to identify USP7-interacting proteins and then combined them with protein-profiling and RNA-seq data. Our assumption was that USP7 substrates may interact with USP7 and be down-regulated in USP7 KO cells, but their mRNA levels should not change in USP7 KO cells. Based on this assumption, we compiled a list of 20 proteins that are likely to be USP7 substrates. This list included some known substrates, such as TRIM27, DHX40, and GMPS, highlighting the quality of our data. Furthermore, we experimentally confirmed MGA and PHIP as novel substrates of USP7. However, although MGA deletion reduced cell proliferation, it could not phenocopy USP7 deletion. Similar results were observed with PHIP knockdown. Indeed, >20 USP7 substrates have been reported (Wang et al. 2019). Moreover, USP7 may function as a context-specific oncogene or tumor suppressor (Li et al. 2004). For example, USP7 can directly destabilize TP53 or indirectly stabilize TP53 by deubiquitinating MDM2 (Li et al. 2002, 2004; Cummins et al. 2004). Of course, our list of 20 high-confidence substrates of USP7 was generated based on data obtained from 293A cells, which by no means is a comprehensive list of potential USP7 substrates. Nevertheless, the two new substrates that we identified in this study (i.e., MGA and PHIP) are also among the top changed proteins in a previous proteomics analysis with a different cell line treated with USP7 inhibitors (XL177A, FT671, and GNE-6640) (Bushman et al. 2021; Steger et al. 2021). We also confirmed the changes of these two proteins in two other cell lines. Thus, it is likely that MGA and PHIP are two major substrates of USP7 in a variety of cellular backgrounds.
USP7 is a promising drug target. Increased TP53 is used as a biomarker to test the specificity of USP7 inhibitors. USP7 inhibitors can boost TP53 level and thus induce apoptosis (Kategaya et al. 2017; Turnbull et al. 2017; Schauer et al. 2020). Sensitivity to USP7 inhibitor is believed to correlate with TP53 mutational status (Schauer et al. 2020). However, USP7 is a pretty promiscuous DUB with many substrates. Even in the TP53 pathway, USP7 can deubiquinate both TP53 and MDM2. Moreover, USP7 has TP53-independent roles, such as destabilizing PTEN and FOXOs (Bhattacharya et al. 2018). Moreover, Tp53 deletion cannot rescue embryonic lethality in Usp7 knockout mice (Kon et al. 2010; Agathanggelou et al. 2017). In this study, we tested two recently developed USP7 inhibitors in three paired wild-type and USP7 KO cells. Unexpectedly, we did not observe any major difference in their sensitivities to these USP7 inhibitors. These data suggest that although these inhibitors could effectively inhibit USP7 activity, they could not provoke strong antiproliferation effects in these three paired wild-type and USP7 KO cells. It is possible that these inhibitors may only be effective in certain genetic and/or tissue contexts, which need to be further defined. Another possibility is that Celltiter-Glo assay, as a short-term assay, cannot reveal the modest difference due to the defective cell proliferation of USP7 KO cells. The long-term colony formation and other assays can be used to further investigate the potential effects of these inhibitors in control and USP7 KO cells.
Although our study systematically revealed the proteome and transcriptome landscape in USP7 KO cells and compiled a list of high-confidence USP7 substrates, there are several limitations in our study. First, it is likely that USP7 may have context-dependent function and substrates. However, our experiments were performed mainly in one cell line (i.e., HEK293A) and under one condition (i.e., with or without USP7 deletion). It is possible that cells may adapt to USP7 KO and therefore preclude the discovery of some USP7 substrates even in these HEK293A cells. Second, potential substrates with low expression abundance and/or moderate change in USP7 KO cells may not be uncovered in our study. Although mass spectrometer technology has advanced rapidly in the past decade, complete coverage and quantification of the whole proteome are still not achievable with the current technology. It is likely that potential substrates with low abundance and/or moderate change were missed in our study. For example, we did not uncover several known USP7 substrates such as c-myc and N-myc (Bhattacharya and Ghosh 2015) in this study. Third, some USP7 substrates may only interact with USP7 weakly and/or transiently. This type of substrate may not be captured by our tandem affinity purification protocol. Fourth, it is possible that some USP7 substrates may be deubiquitinated by USP7, but this regulation does not change their protein levels. Our approach presented here cannot be used to capture these substrates. Despite these limitations, our study is the first proteome-wide profiling of USP7 KO cells. These data, together with RNA-seq and USP7 interactome data, provide a rich resource to facilitate further functional analysis of USP7 and the development of USP7 inhibitors.
Materials and methods
Cell culture
HEK293A, HEK293T, A549, and HeLa cells were purchased from ATCC. HEK293A, HEK293T, and HeLa cells were cultured in Dulbecco's modified Eagle medium (DMEM) plus 10% fetal bovine serum, penicillin, and streptomycin at 37°C in 5% (v/v) CO2. A549 cells were cultured in RPMI 1640 media plus 10% fetal bovine serum and streptomycin at 37°C in 5% (v/v) CO2. CRISPR/Cas9 gene-editing technology was used to generate HEK293A-derived USP7 KO, USP47 KO, and USP7/USP47 DKO clones as described (Nie et al. 2020).
Mass spectrometer analysis
The samples were prepared as described in our previous work (Nie et al. 2020). Briefly, cells were lysed with 8 M urea and subjected to reduction, alkylation, and Trypsin digestion sequentially. After desalination with Sep-Pak column, tryptic peptides were subjected to fractionation as previously described (Nie et al. 2020). The eluent was combined into 10 fractions and then analyzed in a Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific) in data-dependent mode.
Cell proliferation and viability assays
Cells were seeded into 24-well or 96-well plates and counted every day by a Bio-Rad TC20 automated cell counter (Bio-Rad Laboratories) with Trypan Blue solution to confirm the live cells or CellTiter-Glo luminescent cell viability assay with BioTek cytation 5 (BioTek). Growth curves were drawn with three replicates.
For the viability assay following treatment with USP7 inhibitors, cells were plated into 96-well plates at the same density. After 24 h, USP7 inhibitors of the indicated concentrations were added. After 3 d, CellTiter-Glo luminescent cell viability assay was performed following the manufacturer's instructions. Luminescence was measured with BioTek cytation 5 (BioTek) and normalized with DMSO-treated cells. The cell viability curves were drawn with three replicates.
Protein–protein interaction and ubiquitination pull-down assay
For protein–protein interaction, 2 µg of the indicated plasmids (Fig. 5D,G; Supplemental Fig. S7B,D) was transfected into HEK293T cells with 10 µL of polyethylenimines and 200 µL of Opti-MEM (Thermo Fisher Scientific). Approximately 1 × 106 cells were collected 48 h later and lysed with chilled NETN buffer and sonication. The pull-down was performed with the indicated beads for 2 h, beads were washed with NETN three times, and proteins were eluted by boiling for 15 min at 95°C with SDS-PAGE loading buffer for Western blot.
For ubiquitination pull-down, the indicated plasmids were transfected into HEK293T cells. Cells were lysed 48 h later with chilled NETN containing 1% SDS. For denaturation, the lysates were heated for 5 min at 95°C and diluted by 10-fold with lysis buffer (to 0.1% SDS) and sonication. After centrifugation, the lysates were incubated with S-protein beads for 2 h. S-protein beads were washed with lysis buffer three times and subjected to boiling with SDS-PAGE loading buffer for Western blot.
Identification of USP7-associated proteins
To identify USP7-interacting proteins, we followed the tandem affinity purification protocol that we published previously (Chen et al. 2020). Briefly, HEK293T cells with expression N-terminal and SFB-tagged were lysed with chilled NTEN buffer and sonication. After centrifugation, the lysates were incubated with streptavidin-conjugated beads and then eluted with 2 mg/mL Biotin. The elutes were incubated with S-protein beads. The beads were boiled to elute bound proteins with 2× Laemmli sample buffer. Next, the eluted proteins were subjected to in-gel digestion for mass spectrometer analysis.
RNA-seq and data analysis
Cells were processed with RNeasy mini kit (Qiagen 74104) and RNase-free DNase (Qiagen 79254) to extract total RNA according to the manufacturer's instructions. Approximately 10 µg of total RNA was then processed with the Illumina TruSeq stranded total RNA library preparation kit including rRNA depletion, followed by sequencing with NextSeq 500 (Illumina) to generate 75 bp from paired ends.
The raw data were filtered with FastQC. Salmon (version 0.14.1) was used to map reads and assemble transcripts into genes (Patro et al. 2017). The output file from Salmon was processed by DESeq2 (v 1.28.1) to analyze the fold change and adjusted P-values of genes. Genes with adjusted P-value ≤ 0.01 and fold change cutoff of 2 were considered as significantly changed genes.
Statistical analysis and bioinformatics
The proteingroup.txt file was imported into Perseus software (version 1.6.7.0) (Tyanova et al. 2016), followed by filtering proteins annotated with “reverse,” “potential contamination,” and “only identified by site.” Proteins with at least two unique peptides and 70% valid value in total samples were kept. The significantly differential proteins were set as a permutation-based false discovery rate of <0.05 and S0 = 0.1.
The ranked list from protein profiling or RNA-seq was imported into R package clusterProfiler for gene set enrichment analysis (GSEA) (Yu et al. 2012; Isserlin et al. 2013; Wu et al. 2021). The protein–protein interaction network for down-regulated proteins was generated by the STRING database with medium confidence (0.7) and visualized by Cytoscape (version 3.8.2), and the enrichment analysis was conducted by Cytoscape stringApp (Pinkus and Said 1977; Doncheva et al. 2019; Szklarczyk et al. 2019).
Data availability
All proteomics data in this study are presented here and in Supplemental Tables S1–S6. The acquired MS/MS raw data (whole-proteome profiling [Supplemental Table S1] and USP7-associated protein identification [Supplemental Table S5]) in this study have been deposited into MassIVE data sets (ftp://massive.ucsd.edu/MSV000087940). The transcription data were deposited into the GEO database with accession number GSE181513 (password: ilsdgesevhqhzcl).
Supplementary Material
Acknowledgments
We thank all the members of the Chen laboratory for their help and constructive discussions. This work was supported in part by the Pamela and Wayne Garrison Distinguished Chair in Cancer Research to J.C. J.C. also received support from Cancer Prevention and Research Institute of Texas (CPRIT; RP160667 and RP180813) and the National Institutes of Health (NIH; P01CA193124, R01CA210929, R01CA216911, and R01CA216437). This work also received support from CPRIT (RP180734).
Author contributions: L.N., C.W., B.G., L.M., and J.C. conceived the project. L.N., C.W., X.L., H.T., S.L., M.H., X.F., G.P., Q.H., and Z.Z. performed the experiments and analyzed the data. L.N. and J.C. wrote the manuscript with input from all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349848.122.
Competing interest statement
The authors declare no competing interests.
References
- Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K, Hofmann K, Kulathu Y. 2016. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell 63: 146–155. 10.1016/j.molcel.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agathanggelou A, Smith E, Davies NJ, Kwok M, Zlatanou A, Oldreive CE, Mao J, Da Costa D, Yadollahi S, Perry T. 2017. USP7 inhibition alters homologous recombination repair and targets CLL cells independently of ATM/p53 functional status. Blood 130: 156–166. [DOI] [PubMed] [Google Scholar]
- Becker K, Marchenko ND, Palacios G, Moll UM. 2008. A role of HAUSP in tumor suppression in a human colon carcinoma xenograft model. Cell Cycle 7: 1205–1213. 10.4161/cc.7.9.5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Ghosh MK. 2015. HAUSP regulates c-MYC expression via de-ubiquitination of TRRAP. Cell Oncol 38: 265–277. 10.1007/s13402-015-0228-6 [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Chakraborty D, Basu M, Ghosh MK. 2018. Emerging insights into HAUSP (USP7) in physiology, cancer and other diseases. Signal Transduct Target Ther 3: 17. 10.1038/s41392-018-0012-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman JW, Donovan KA, Schauer NJ, Liu X, Hu W, Varca AC, Buhrlage SJ, Fischer ES. 2021. Proteomics-based identification of DUB substrates using selective inhibitors. Cell Chem Biol 28: 78–87.e3. 10.1016/j.chembiol.2020.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Chen HY, Peng SJ, Meng JL, Wang Y, Zhou Y, Qian XP, Sun XY, Pang XW, Zhang Y, et al. 2018. USP7–TRIM27 axis negatively modulates antiviral type I IFN signaling. FASEB J 32: 5238–5249. 10.1096/fj.201700473RR [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2014. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511: 543–550. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrà G, Panuzzo C, Torti D, Parvis G, Crivellaro S, Familiari U, Volante M, Morena D, Lingua MF, Brancaccio M, et al. 2017. Therapeutic inhibition of USP7–PTEN network in chronic lymphocytic leukemia: a strategy to overcome TP53 mutated/deleted clones. Oncotarget 8: 35508–35522. 10.18632/oncotarget.16348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catic A, Fiebiger E, Korbel GA, Blom D, Galardy PJ, Ploegh HL. 2007. Screen for ISG15-crossreactive deubiquitinases. PLoS One 2: e679. 10.1371/journal.pone.0000679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wang C, Jain A, Srivastava M, Tang M, Zhang H, Feng X, Nie L, Su D, Xiong Y, et al. 2020. AMPK interactome reveals new function in non-homologous end joining DNA repair. Mol Cell Proteomics 19: 467–477. 10.1074/mcp.RA119.001794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Niu C, Yang Y, Wang Y, Lu M. 2013. Expression of HAUSP in gliomas correlates with disease progression and survival of patients. Oncol Rep 29: 1730–1736. 10.3892/or.2013.2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Yang H, Fang J, Ma L, Gong R, Wang P, Li Z, Xu Y. 2015. Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation. Nat Commun 6: 7023. 10.1038/ncomms8023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. 1998. The ubiquitin–proteasome pathway: on protein death and cell life. EMBO J 17: 7151–7160. 10.1093/emboj/17.24.7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleran A, Collins PE, O'Carroll C, Ahmed A, Mao X, McManus B, Kiely PA, Burstein E, Carmody RJ. 2013. Deubiquitination of NF-κB by ubiquitin-specific protease-7 promotes transcription. Proc Natl Acad Sci 110: 618–623. 10.1073/pnas.1208446110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. 2004. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature 428: 1 p following 486. 10.1038/nature02501 [DOI] [PubMed] [Google Scholar]
- D'Arcy P, Wang X, Linder S. 2015. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther 147: 32–54. 10.1016/j.pharmthera.2014.11.002 [DOI] [PubMed] [Google Scholar]
- Daubeuf S, Singh D, Tan Y, Liu H, Federoff HJ, Bowers WJ, Tolba K. 2009. HSV ICP0 recruits USP7 to modulate TLR-mediated innate response. Blood 113: 3264–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paoli L, Cerri M, Monti S, Rasi S, Spina V, Bruscaggin A, Greco M, Ciardullo C, Famà R, Cresta S, et al. 2013. MGA, a suppressor of MYC, is recurrently inactivated in high risk chronic lymphocytic leukemia. Leuk Lymphoma 54: 1087–1090. 10.3109/10428194.2012.723706 [DOI] [PubMed] [Google Scholar]
- Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. 2019. Cytoscape stringApp: network analysis and visualization of proteomics data. J Proteome Res 18: 623–632. 10.1021/acs.jproteome.8b00702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J 16: 1519–1530. 10.1093/emboj/16.7.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. 2011. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell 44: 147–159. 10.1016/j.molcel.2011.06.034 [DOI] [PubMed] [Google Scholar]
- Fountain MD, Oleson DS, Rech ME, Segebrecht L, Hunter JV, McCarthy JM, Lupo PJ, Holtgrewe M, Moran R, Rosenfeld JA, et al. 2019. Pathogenic variants in USP7 cause a neurodevelopmental disorder with speech delays, altered behavior, and neurologic anomalies. Genet Med 21: 1797–1807. 10.1038/s41436-019-0433-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges A, Marcon E, Greenblatt J, Frappier L. 2018. Identification and characterization of USP7 targets in cancer cells. Sci Rep 8: 15833. 10.1038/s41598-018-34197-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges A, Coyaud E, Marcon E, Greenblatt J, Raught B, Frappier L. 2019. USP7 regulates cytokinesis through FBXO38 and KIF20B. Sci Rep 9: 2724. 10.1038/s41598-019-39368-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. 1998. The ubiquitin system. Annu Rev Biochem 67: 425–479. 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L. 2003a. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J Biol Chem 278: 47753–47761. 10.1074/jbc.M307200200 [DOI] [PubMed] [Google Scholar]
- Holowaty MN, Zeghouf M, Wu H, Tellam J, Athanasopoulos V, Greenblatt J, Frappier L. 2003b. Protein profiling with Epstein–Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J Biol Chem 278: 29987–29994. 10.1074/jbc.M303977200 [DOI] [PubMed] [Google Scholar]
- Hurlin PJ, Steingrimsson E, Copeland NG, Jenkins NA, Eisenman RN. 1999. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J 18: 7019–7028. 10.1093/emboj/18.24.7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson MK, Ploegh HL. 2009. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5: 559–570. 10.1016/j.chom.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isserlin R, Merico D, Emili A. 2013. Global proteomic profiling and enrichment maps of dilated cardiomyopathy. Methods Mol Biol 1005: 53–66. 10.1007/978-1-62703-386-2_5 [DOI] [PubMed] [Google Scholar]
- Jang SM, Zhang Y, Utani K, Fu H, Redon CE, Marks AB, Smith OK, Redmond CJ, Baris AM, Tulchinsky DA, et al. 2018. The replication initiation determinant protein (RepID) modulates replication by recruiting CUL4 to chromatin. Nat Commun 9: 2782. 10.1038/s41467-018-05177-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Martinez CA, Arcipowski KM, Zhu Y, Gutierrez-Diaz BT, Wang KK, Johnson MR, Volk AG, Wang F, Wu J, et al. 2019. USP7 cooperates with NOTCH1 to drive the oncogenic transcriptional program in T-cell leukemia. Clin Cancer Res 25: 222–239. 10.1158/1078-0432.CCR-18-1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kategaya L, Di Lello P, Rougé L, Pastor R, Clark KR, Drummond J, Kleinheinz T, Lin E, Upton JP, Prakash S, et al. 2017. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 550: 534–538. 10.1038/nature24006 [DOI] [PubMed] [Google Scholar]
- Kim RQ, Sixma TK. 2017. Regulation of USP7: a high incidence of E3 complexes. J Mol Biol 429: 3395–3408. 10.1016/j.jmb.2017.05.028 [DOI] [PubMed] [Google Scholar]
- Kon N, Kobayashi Y, Li M, Brooks C, Ludwig T, Gu W. 2010. Inactivation of HAUSP in vivo modulates p53 function. Oncogene 29: 1270–1279. 10.1038/onc.2009.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasna D, Abdul Rehman SA, Natarajan J, Matthews S, Madden R, De Cesare V, Weidlich S, Virdee S, Ahel I, Gibbs-Seymour I, et al. 2018. Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability. Mol Cell 70: 150–164.e6. 10.1016/j.molcel.2018.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SM, Armstrong LA, Kulathu Y. 2022. Deubiquitinases: from mechanisms to their inhibition by small molecules. Mol Cell 82: 15–29. 10.1016/j.molcel.2021.10.027 [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416: 648–653. 10.1038/nature737 [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, Gu W. 2004. A dynamic role of HAUSP in the p53–Mdm2 pathway. Mol Cell 13: 879–886. 10.1016/S1097-2765(04)00157-1 [DOI] [PubMed] [Google Scholar]
- Liu Q, Wu Y, Qin Y, Hu J, Xie W, Qin FX, Cui J. 2018. Broad and diverse mechanisms used by deubiquitinase family members in regulating the type I interferon signaling pathway during antiviral responses. Sci Adv 4: eaar2824. 10.1126/sciadv.aar2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhao H, Yu C, Kang Y, Yang X. 2021. Targeting ubiquitin-specific protease 7 (USP7) in cancer: a new insight to overcome drug resistance. Front Pharmacol 12: 648491. 10.3389/fphar.2021.648491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maat H, Atsma TJ, Hogeling SM, Rodríguez López A, Jaques J, Olthuis M, de Vries MP, Gravesteijn C, Brouwers-Vos AZ, van der Meer N, et al. 2021. The USP7–TRIM27 axis mediates non-canonical PRC1.1 function and is a druggable target in leukemia. iScience 24: 102435. 10.1016/j.isci.2021.102435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MAJ, Rickels RA, Collings CK, He X, Cao K, Herz HM, Cozzolino KA, Abshiru NA, Marshall SA, Rendleman EJ, et al. 2017. A cryptic Tudor domain links BRWD2/PHIP to COMPASS-mediated histone H3K4 methylation. Genes Dev 31: 2003–2014. 10.1101/gad.305201.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L, Wang C, Li N, Feng X, Lee N, Su D, Tang M, Yao F, Chen J. 2020. Proteome-wide analysis reveals substrates of E3 ligase RNF146 targeted for degradation. Mol Cell Proteomics 19: 2015–2030. 10.1074/mcp.RA120.002290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazón-Riquelme P, Worboys JD, Green J, Valera A, Martín-Sánchez F, Pellegrini C, Brough D, López-Castejón G. 2018. USP7 and USP47 deubiquitinases regulate NLRP3 inflammasome activation. EMBO Rep 19: e44766. 10.15252/embr.201744766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JL, Dianova II, Khoronenkova SV, Edelmann MJ, Kessler BM, Dianov GL. 2011. USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase β. Mol Cell 41: 609–615. 10.1016/j.molcel.2011.02.016 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Liu Y, Gao Y, Yuan B, Qi X, Fu Y, Zhu Q, Cao T, Zhang S, Yin L, et al. 2019. USP7 is a novel deubiquitinase sustaining PLK1 protein stability and regulating chromosome alignment in mitosis. J Exp Clin Cancer Res 38: 468. 10.1186/s13046-019-1457-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfoh R, Lacdao IK, Georges AA, Capar A, Zheng H, Frappier L, Saridakis V. 2015. Crystal structure of USP7 ubiquitin-like domains with an ICP0 peptide reveals a novel mechanism used by viral and cellular proteins to target USP7. PLoS Pathog 11: e1004950. 10.1371/journal.ppat.1004950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao J, Tashiro A, Nishikawa M, Aoki Y, Moriyoshi E, Hattori A, Kakeya H. 2015. Expression, purification and enzymatic characterization of a recombinant human ubiquitin-specific protease 47. J Biochem 158: 477–484. [DOI] [PubMed] [Google Scholar]
- Pinkus GS, Said JW. 1977. Specific identification of intracellular immunoglobulin in paraffin sections of multiple myeloma and macroglobulinemia using an immunoperoxidase technique. Am J Pathol 87: 47–57. [PMC free article] [PubMed] [Google Scholar]
- Pozhidaeva A, Bezsonova I. 2019. USP7: structure, substrate specificity, and inhibition. DNA Repair 76: 30–39. 10.1016/j.dnarep.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Wang C, Zhu Y, Su T, Dong L, Huang Y, Hao K. 2021. Mga safeguards embryonic stem cells from acquiring extraembryonic endoderm fates. Sci Adv 7: eabe5689. 10.1126/sciadv.abe5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougé L, Bainbridge TW, Kwok M, Tong R, Di Lello P, Wertz IE, Maurer T, Ernst JA, Murray J. 2016. Molecular understanding of USP7 substrate recognition and C-terminal activation. Structure 24: 1335–1345. 10.1016/j.str.2016.05.020 [DOI] [PubMed] [Google Scholar]
- Schauer NJ, Liu X, Magin RS, Doherty LM, Chan WC, Ficarro SB, Hu W, Roberts RM, Iacob RE, Stolte B, et al. 2020. Selective USP7 inhibition elicits cancer cell killing through a p53-dependent mechanism. Sci Rep 10: 5324. 10.1038/s41598-020-62076-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft D, Qi J, Ronai ZA. 2018. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer 18: 69–88. 10.1038/nrc.2017.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H, Li X, Xiao X, Dai Y, Huang J, Song J, Liu M, Yang L, Lei H, Tong Y, et al. 2018. USP7 deubiquitinates and stabilizes NOTCH1 in T-cell acute lymphoblastic leukemia. Signal Transduct Target Ther 3: 29. 10.1038/s41392-018-0028-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, Frappier L. 2006. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol 13: 285–291. 10.1038/nsmb1067 [DOI] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. 2008. The deubiquitinylation and localization of PTEN are regulated by a HAUSP–PML network. Nature 455: 813–817. 10.1038/nature07290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Chen Z, Zhang H, Tang M, Wang C, Jung SY, Chen J. 2018. Replisome dynamics and their functional relevance upon DNA damage through the PCNA interactome. Cell Rep 25: 3869–3883.e4. 10.1016/j.celrep.2018.11.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger M, Demichev V, Backman M, Ohmayer U, Ihmor P, Müller S, Ralser M, Daub H. 2021. Time-resolved in vivo ubiquitinome profiling by DIA-MS reveals USP7 targets on a proteome-wide scale. Nat Commun 12: 5399. 10.1038/s41467-021-25454-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stielow B, Finkernagel F, Stiewe T, Nist A, Suske G. 2018. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet 14: e1007193. 10.1371/journal.pgen.1007193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Wang W, Hou Y, Wang L, Yi X, Cao C, Wang Y, Gao H, Wang Y, Yang C, et al. 2021. Bimodal regulation of the PRC2 complex by USP7 underlies tumorigenesis. Nucleic Acids Res 49: 4421–4440. 10.1093/nar/gkab209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. 2008. Deubiquitylation and regulation of the immune response. Nat Rev Immunol 8: 501–511. 10.1038/nri2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. 2019. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47: D607–D613. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Sun X, Hou F-Y, Oh H, Hilgenberg L, Hol E, Van Leeuwen F, Smith M, O'Dowd DK, Schreiber S. 2007. Mutant ubiquitin found in Alzheimer's disease causes neuritic beading of mitochondria in association with neuronal degeneration. Cell Death Differ 14: 1721–1732. 10.1038/sj.cdd.4402180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Isamiddinova NS, Peroutka RJ, Goldenberg SJ, Mattern MR, Nicholson B, Leach C. 2011. Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format. Assay Drug Dev Technol 9: 165–173. 10.1089/adt.2010.0317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull AP, Ioannidis S, Krajewski WW, Pinto-Fernandez A, Heride C, Martin ACL, Tonkin LM, Townsend EC, Buker SM, Lancia DR, et al. 2017. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 550: 481–486. 10.1038/nature24451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Cox J. 2018. Perseus: a bioinformatics platform for integrative analysis of proteomics data in cancer research. Methods Mol Biol 1711: 133–148. 10.1007/978-1-4939-7493-1_7 [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- van Loosdregt J, Fleskens V, Fu J, Brenkman AB, Bekker CP, Pals CE, Meerding J, Berkers CR, Barbi J, Grone A, et al. 2013. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg cell-suppressive capacity. Immunity 39: 259–271. 10.1016/j.immuni.2013.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Ma S, Song N, Li X, Liu L, Yang S, Ding X, Shan L, Zhou X, Su D, et al. 2016. Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J Clin Invest 126: 2205–2220. 10.1172/JCI85747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kang W, You Y, Pang J, Ren H, Suo Z, Liu H, Zheng Y. 2019. USP7: novel drug target in cancer therapy. Front Pharmacol 10: 427. 10.3389/fphar.2019.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washkowitz AJ, Schall C, Zhang K, Wurst W, Floss T, Mager J, Papaioannou VE. 2015. Mga is essential for the survival of pluripotent cells during peri-implantation development. Development 142: 31–40. 10.1242/dev.111104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock J, Wu J, Cao P, Kingsbury WD, McDermott JL, Kodrasov MP, McKelvey DM, Suresh Kumar KG, Goldenberg SJ, Mattern MR, et al. 2012. Selective dual inhibitors of the cancer-related deubiquitylating proteases USP7 and USP47. ACS Med Chem Lett 3: 789–792. 10.1021/ml200276j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. 2021. Clusterprofiler 4.0: a universal enrichment tool for interpreting omics data. The Innovation 2: 100141. 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. 2012. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZM, Rothbart SB, Allison DF, Cai Q, Harrison JS, Li L, Wang Y, Strahl BD, Wang GG, Song J. 2015. An allosteric interaction links USP7 to deubiquitination and chromatin targeting of UHRF1. Cell Rep 12: 1400–1406. 10.1016/j.celrep.2015.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All proteomics data in this study are presented here and in Supplemental Tables S1–S6. The acquired MS/MS raw data (whole-proteome profiling [Supplemental Table S1] and USP7-associated protein identification [Supplemental Table S5]) in this study have been deposited into MassIVE data sets (ftp://massive.ucsd.edu/MSV000087940). The transcription data were deposited into the GEO database with accession number GSE181513 (password: ilsdgesevhqhzcl).