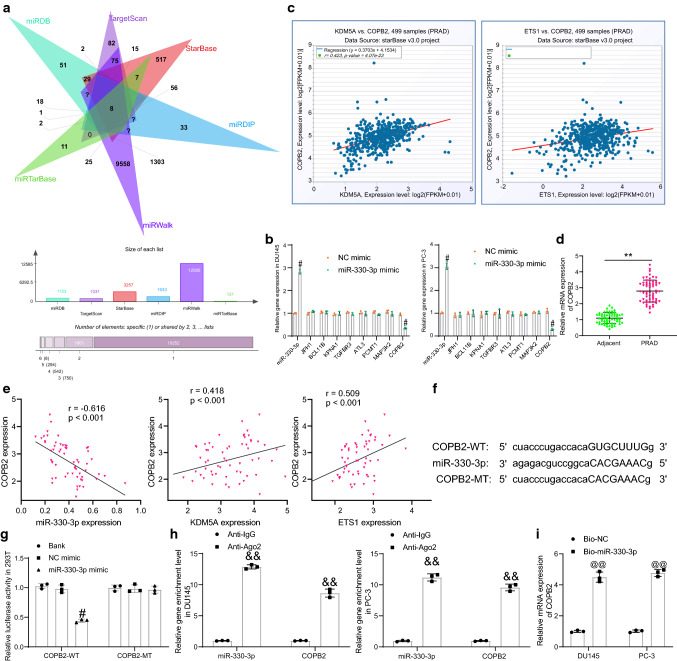
Fig. 6.
miR-330-3p targets COPB2. A candidate target mRNAs of miR-330-3p predicted using six bioinformatic systems; B expression of miR-330-3p and COPB2 in DU145 and PC-3 cells after miR-330-3p mimic transfection determined by RT-qPCR; C correlations between each COPB2 with ETS1 and KDM5A in PRAD analyzed on StarBase; D COPB2 expression in the collected PRAD tumor tissues and the adjacent tissues evaluated by RT-qPCR (n = 60); E correlations between COPB2 expression and miR-330-3p, ETS1 and KDM5A expression in the collected PRAD tumor tissues; F sequences of COPB2-WT/COPB2-MT vectors for luciferase assay; G binding relationship between miR-330-3p and COPB2 in cells examined by a dual luciferase reporter gene assay; H enrichment of miR-330-3p and COPB2 fragments by anti-Ago2 in RIP assay; I enrichment of COPB2 fragments by Bio-miR-330-3p examined by RNA pull-down assay. Data were collected from three independent experiments and expressed as mean ± SEM. Differences were analyzed by paired t test (D) or two-way ANOVA (B, G and H); correlations between variables (E) were analyzed by Pearson’s correlation analysis (COPB2 vs. miR-330-3p: r = -0.616, p < 0.001; COPB2 vs. KDM5A; r = 0.418, p < 0.01; COPB2 vs. ETS1: r = 0.509, p < 0.001); **p < 0.01 compared to adjacent tissues; #p < 0.05 NC mimic; &&p < 0.01 compared to anti-IgG; @@p < 0.01 compared to Bio-NC