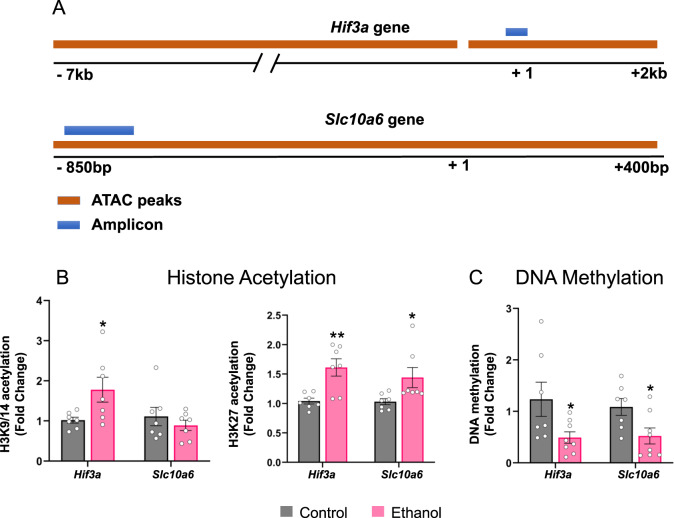
Fig. 3. Histone acetylation (H3K9/14ac; H3K27ac) and DNA methylation (5-methylcytosine, 5mC) changes reveal relaxed chromatin architecture at the ATAC-seq peak regions near the transcription start site (TSS) of Hif3a and Slc10a6 genes.
A Schematic showing the location of ATAC-seq peaks and location of primers for qPCR validation for Hif3a and Slc10a6. B The “open peak” regions at the transcriptional control regions of Hif3a and Slc10a6 were evaluated by chromatin immunoprecipitation (ChIP) assay representing fold changes of H3K27ac and H3K9/14ac occupancy at selected regions of Hif3a and Slc10a6 in ethanol-treated and control rats. C Changes in DNA methylation were also evaluated at the same loci of Hif3a and Slc10a6 in amygdala of ethanol-treated and control rats and represented as fold changes in DNA methylation (5mC levels). Values are represented as the mean ± SEM and individual values are shown on bar diagrams with circle dots for control and ethanol groups (n = 7–8; two-tailed Student’s t test *p < 0.05; **p < 0.01).