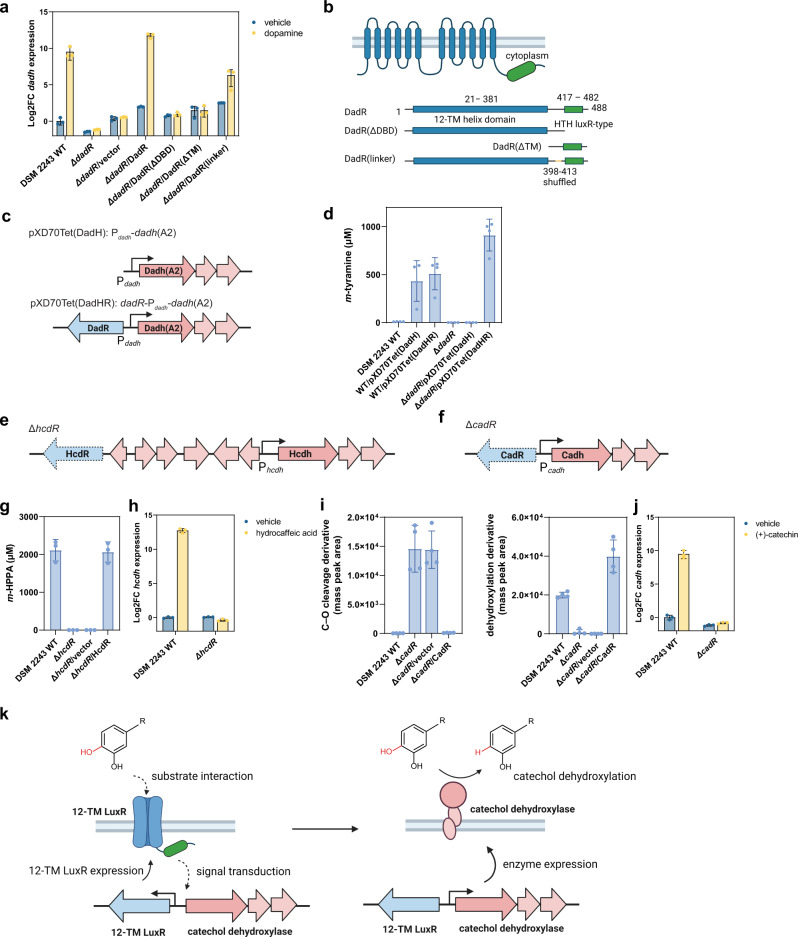
Fig. 6. Gene deletion and complementation reveal that E. lenta DadR, HcdR, and CadR are transmembrane transcriptional activators.
a RT-qPCR to test dadh upregulation on exposure to dopamine in E. lenta DSM 2243 WT, ΔdadR and complementation strains. b Schematic of the domain organization of DadR and DadR mutants examined in this work. c Schematic of pXD70Tet(DadH) and pXD70Tet(DadHR) constructs for testing the relevance of DadR for E. lenta dopamine metabolism. d LC-MS/MS to quantify the production of dopamine dehydroxylation metabolite m-tyramine after incubation with corresponding E. lenta cultures for 72 h. e Deletion of hcdR in E. lenta DSM 2243 to generate ΔhcdR strain. f Deletion of cadR in E. lenta DSM 2243 to generate ΔcadR strain. g LC-MS/MS to quantify the production of hydrocaffeic acid dehydroxylation metabolite m-HPPA after incubation with corresponding E. lenta cultures for 48 h. h RT-qPCR to test hcdh upregulation on exposure to hydrocaffeic acid in E. lenta DSM 2243 WT and ΔhcdR. i, LC-MS/MS to quantify the production of (+)-catechin C–O cleavage metabolite and dehydroxylation metabolite after incubation with corresponding E. lenta cultures for 48 h. j RT-qPCR to test cadh upregulation on exposure to (+)-catechin in E. lenta DSM 2243 WT and ΔcadR. k Proposed mechanisms of 12-TM LuxR-mediated gene regulation in response to catechol inducer. Data represented as mean ± S.E.M. with n = 3 biological replicates in a, h, and j. Data represented as mean ± SD with n = 4 biological replicates in d and i. Data represented as mean ± SD with n = 3 biological replicates in g. Source data are provided as a Source Data file.