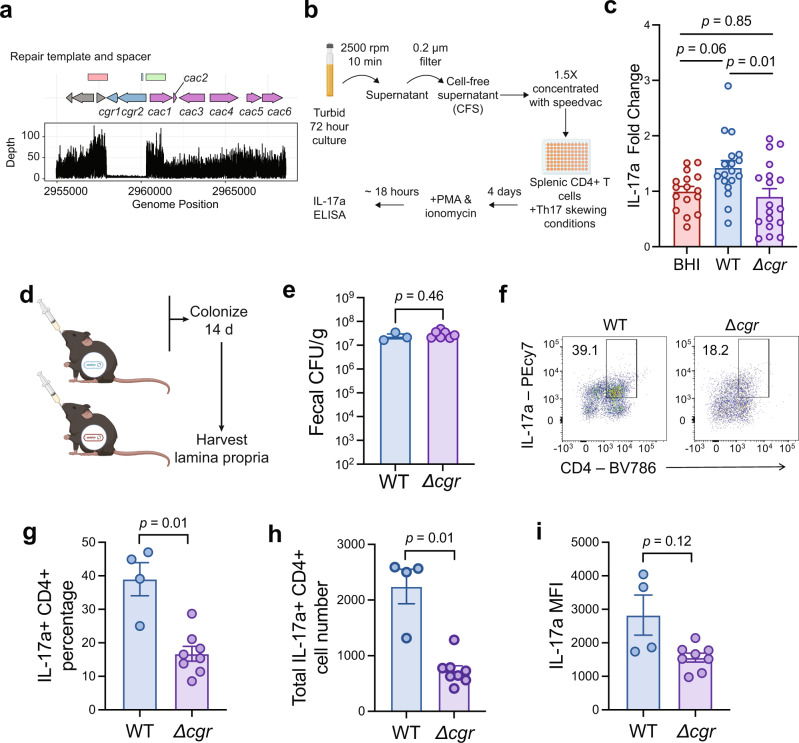
Fig. 8. Genomic engineering and in vivo experiment show that the cgr operon is necessary for colonic Th17 induction by E. lenta.
a Whole genome sequencing confirms the targeted deletion of the cgr operon. Additional reads flanking the deletion site are due to the presence of the editing plasmid in the long-read sequencing run, as indicated by red and green rectangles. b Schematic of in vitro Th17 skewing assay. c Fold-change of IL-17a levels relative to BHI control of CFS from E. lenta WT or Δcgr. d Experimental outline of mouse experiment. Germ-free C57BL/6J male mice ages 6–8 weeks were separated into groups and gavaged with E. lenta WT (n = 4) and Δcgr (n = 8). Bacteria were allowed to colonize for 2 weeks before the lamina propria was harvested. e No significant differences in E. lenta colonization on day 14. f Representative flow cytometry plots of colonic CD4+ IL-17a+ cells within the live CD3+ gate. Percentage of CD4 + IL-17a+ cells is displayed on the flow plot. g Percentage of colonic IL-17a+ CD4+ cells within the live CD3+ gate. h Total numbers of colonic IL-17a+ CD4+ cells within the live CD3+ gate. i Mean fluorescence intensity (MFI) of colonic IL-17a. All p-values are displayed and were calculated with one-way ANOVA tests with Tukey’s multiple correction or Welch’s t tests for two-way comparisons. Data represented as mean ± SD in c (BHI n = 16, WT and Δcgr n = 18 biological replicates), e (WT n = 3, Δcgr n = 7 because fecal pellets were unable to be collected from mouse 4 of the E. lenta WT group and mouse 8 of the Δcgr group) and g–i (WT n = 4, Δcgr n = 8). Each dot represents an individual biological replicate in c or an individual mouse in e, g, h, and i. Panels e–i show representative data from the first of two independent experiments. Source data are provided as a Source Data file.