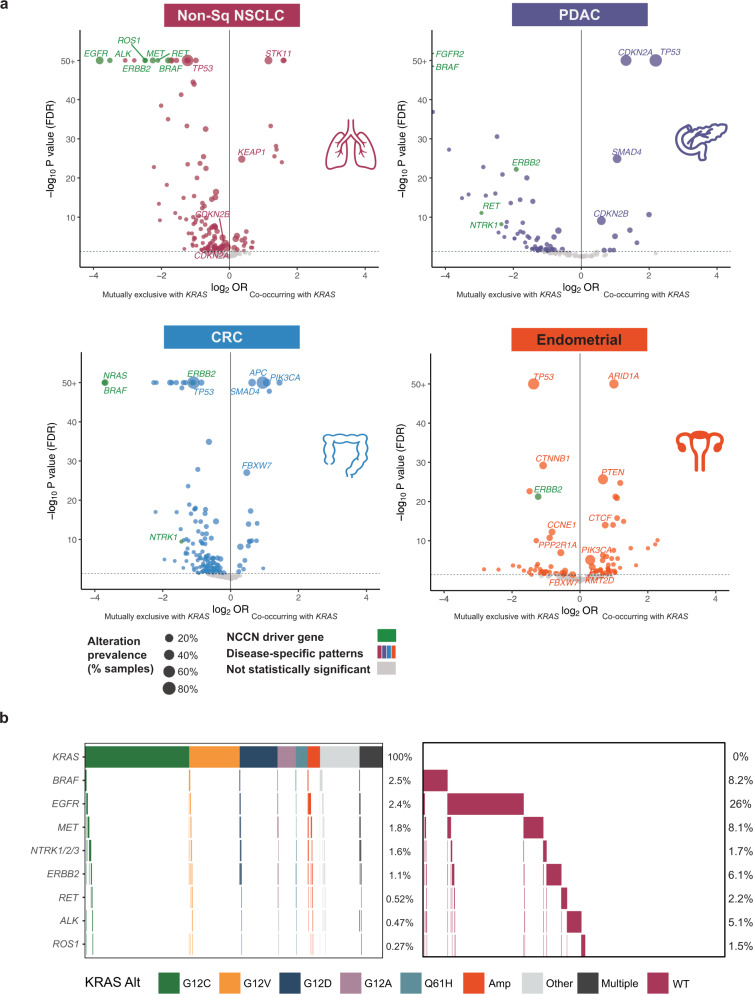
Fig. 3. Co-occurrence of gene alterations among KRAS altered non-Sq NSCLC, PDAC, CRC and endometrial cancer.
a The prevalence of alterations was compared for KRAS altered and KRAS wild type (WT) non-Sq NSCLC (N = 62,836), PDAC (N = 19,386), CRC (N = 48,905), and endometrial (N = 14,375) tumor samples. For each tumor cohort, only genes altered in at least 50 cases and targeted across all the assay versions were included. For each gene, substitutions, short insertions/deletions, rearrangements, and copy number changes of known or likely functional significance detected using our assay were included. Driver genes highlighted in the National Comprehensive Cancer Network (NCCN) Guidelines as well as genes altered at a high prevalence (≥10%) are labeled for each volcano plot. Alterations in known driver oncogenes (labeled in green) tend to be mutually exclusive with KRAS alterations (left side of plots) in all four major tumor types studied (p ≤ 0.05; Odd’s ratio <1). b Oncoprints showing the frequency and mutual exclusivity of NCCN driver genes in KRAS altered (N = 22,794) vs KRAS WT (N = 40,042) non-Sq NSCLC. Fisher’s exact test was applied to assess patterns of co-occurrence and mutual exclusivity between KRAS and other genes alterations. P values were corrected with the Benjamini–Hochberg FDR method.