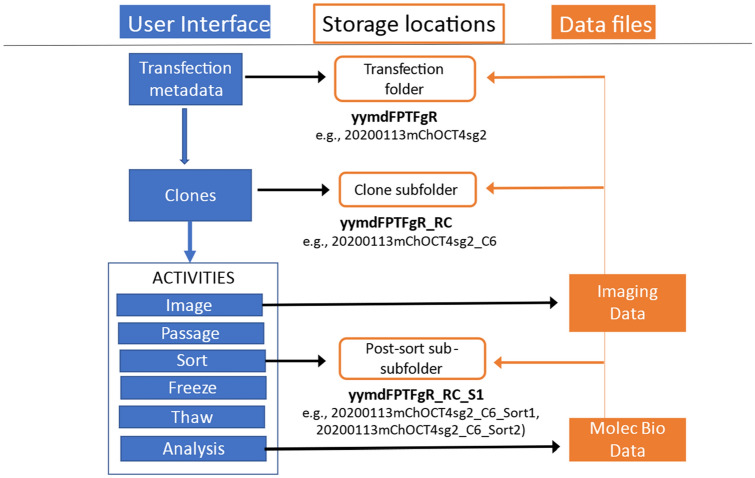
Figure 3.
Schematic of the data model and naming convention. Various activities were performed on each cell sample. Initiating a transfection resulting in generating a unique transfection designator, or cell sample name, which served as the base name for subsequently derived cell samples. Updated cell sample names were created by appending terms to the transfection designator to indicate activities performed on the sample. Using a cell sample-naming convention, storage location folders were automatically generated with those unique sample designators, providing a local persistent identifier and an organizational structure for storing related sample-specific data. In this naming convention, FP represents fluorescent protein, TF represents transcription factor, and gR represents guide RNA. Row “R” and column “C” locations in 96-well plates where edited cells were observed (e.g. “_C6”) provided an appended designator for that cell sample that was isolated and further acted on. Flow cytometry sorting was used to enrich and expand populations of edited cells, which were indicated by appending to the sample name the designation “_ Sort#”. The “#” was incremented automatically for samples as they were subjected to subsequent sorting events.