Figure 1.
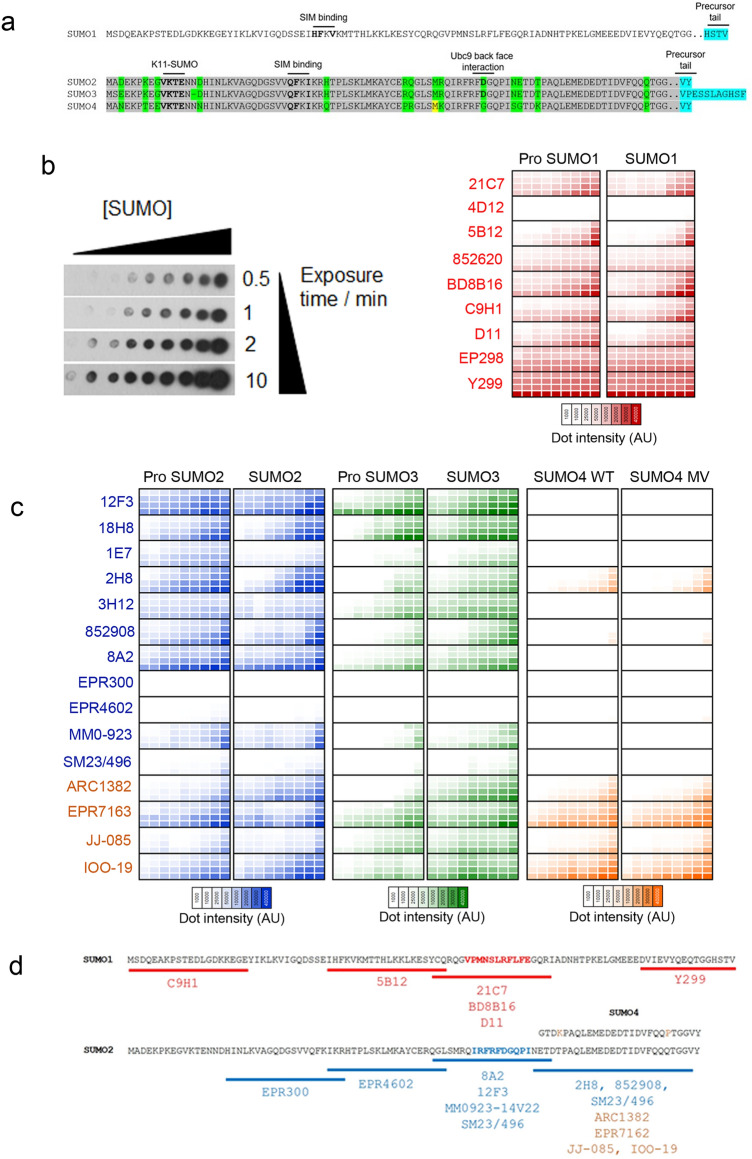
Variable sensitivity and selectivity of SUMO MAbs to detect monomeric SUMO. (a) Illustration of SUMO1-4 sequence. Amino acid sequences of human SUMO1 (P63165), SUMO2 (P61956), SUMO3 (P55854) and SUMO4 (Q6EEV6). The major SUMO acceptor K11, SIM (SUMO Interacting Motif) contacting residues, Ubc9 back face interacting residues are highlighted in bold. Precursor tail residues cleaved by SENP enzymes are indicated in blue. Conserved residues between SUMO2-4 are highlighted in grey, while non-conserved residues are highlighted in green. The variable M55 residue in SUMO4 is highlighted in yellow. (b) Right—heat map of SUMO1 MAbs detecting pro and mature rSUMO1. rSUMO1 was spotted (1 μL) at a concentration of 3 μg/μL with half dilutions per spot down to 3 ng/μL. A deeper colour signifies a more intense chemiluminescent signal, for four exposure time points (0.5, 1, 2 and 10 min—shown vertically) as a mean of three independent repeats based on densitometry of chemiluminescent detection on X-ray film. Left -a representative dot blot is shown with increasing concentration of rSUMO1 on the horizontal and increasing film exposure time on the vertical. N = 3. (c) Heat map of SUMO2/3/4 MAbs detecting pro and mature rSUMO2/3/4. As for 1b, in this case, each MAb was tested against SUMO2 and SUMO3 (Pro and mature forms) and SUMO4 (WT and M55V variant). SUMO2/3/4 were diluted at the same concentrations as rSUMO1. SUMO2/3 MAbs are in blue text and SUMO4 MAbs are in orange. N = 3. (d) Epitope mapping of SUMO MAbs. Partially overlapping SUMO1, SUMO2 or SUMO4 peptides, shown by underline, were used for competition assays. Loss of detection (by dot blot or immunoblot of SUMO conjugates) was interpreted as indicating the location of individual MAb epitopes. The SUMO1 region highlighted in red shows the mapped 21C7 epitope. The SUMO2 region highlighted in blue shows the 8A2 epitope. SUMO4 MAbs are shown in orange. The sequence of SUMO4 is shown above, with the two non-conserved residues highlighted in orange. MAbs not shown could not be mapped.