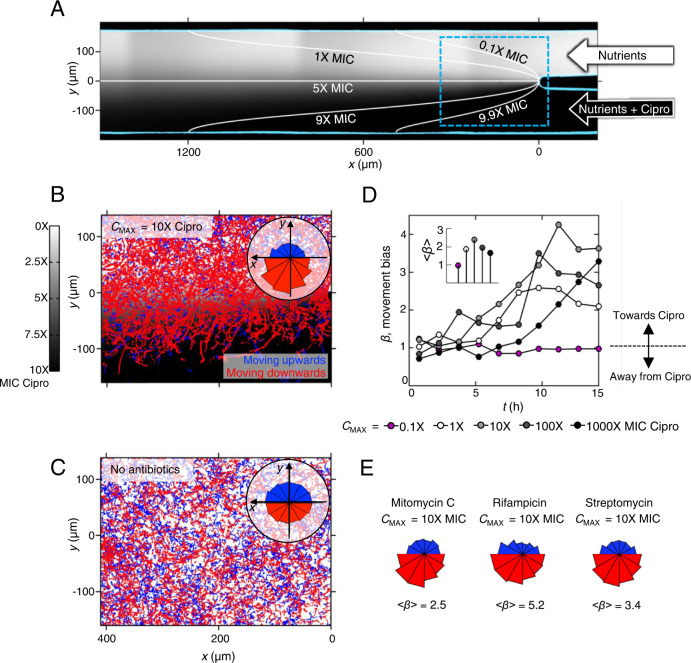
Fig. 1. Twitching P. aeruginosa cells bias their motility towards increasing antibiotic concentrations.
A A dual-inlet microfluidic device generates steady antibiotic gradients (e.g. ciprofloxacin, CMAX = 10X MIC) via molecular diffusion. Isocontours were calculated using mathematical modelling (Methods) and background shading shows approximate ciprofloxacin distribution visualised using fluorescein. B Red (blue) cell trajectories are moving towards (away from) increasing [ciprofloxacin]. Inset: A circular histogram of cell movement direction reveals movement bias towards increasing [ciprofloxacin]. A two-sided binomial test rejects the null hypothesis that trajectories are equally likely to be directed up or down the [ciprofloxacin] gradient (p < 0.0001, n = 10,714 trajectories). C No such bias is seen when nutrient medium without ciprofloxacin is added to both inlets (p = 0.854, n = 8138). D The movement bias, β , is the number of cells moving up divided by the number moving down the gradient. Cell movement is initially (t < ≈5 h) unbiased (β ≈ 1), after which cells bias their movement towards ciprofloxacin (β > 1), even when CMAX = 1000X MIC, (black circles). Movement remains unbiased when CMAX = 0.1X MIC, (magenta circles). At t = 15 h, a two-sided binomial test accepts the null hypothesis that trajectories are equally likely to be directed up or down the [ciprofloxacin] gradient for CMAX = 0.1X MIC (p = 0.811, n = 625 cell trajectories) and rejects it for CMAX = 1X, 10X, 100X and 1000X (p < 0.0001 for each, n = 653, 934, 642 and 589 respectively). Inset: The overall bias calculated across all time, <β > , shows that CMAX = 10X MIC induces the largest response; a two-sided chi-squared test that compared the number of trajectories moving up or down the gradient showed that CMAX = 10X MIC produced a statistically distinct response from the other CMAX values, (p < 0.0001, n = 13,370 cell trajectories across all data sets). E Cells also bias movement towards other antibiotics (p < 0.0001, n = 1778, 1673 and 3347 trajectories from left-to-right). Figures S2 and S5 show biological repeats and source data are provided as a Source Data file.