Figure 2.
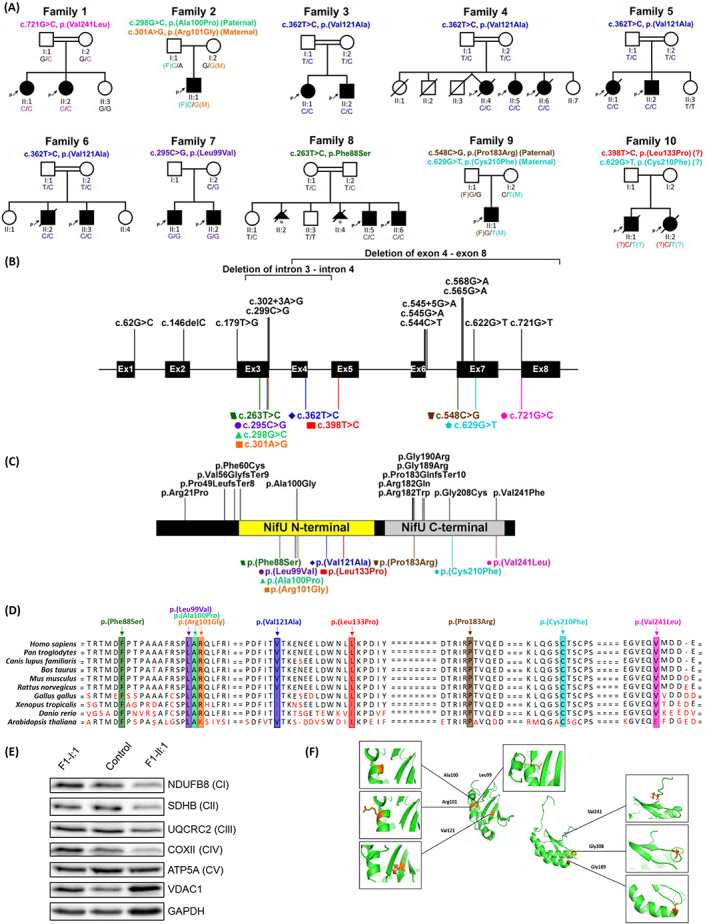
Overview of the genetic and biochemical characteristics of NFU1 variants. (A) Pedigrees and segregation results of the ten families included in this study. (B) NFU1 gene structure with the localization of all previously known mutations (black, above) and mutations reported in this cohort (colored, below). The corresponding amino acid changes is shown in (C) with known protein domains in NFU1 (NifU N‐terminal domain, residue 59 to 155, and NifU C‐terminal domain, residue 162 to 247) depicted in the illustration. The variant, p.(Val241Leu), was the only mutation discovered in this study to have been located outside of the NFU1 protein domains. Regions are not drawn to scales and both illustrations were created from the program. 16 (D) Conservation of the nine mutations reported in the present cohort across 10 species. (E) Western blot analysis of structural subunits from each OXPHOS complex (CI [NDUFB8], CII [SDHB], CIII [UQCRC2], CIV [COXII], and CV [ATP5A]) in fibroblasts from F1‐II:1 (affected individual), F1‐I:1 (mother) and a pediatric control. GAPDH and VDAC1 were used as cell and mitochondrial loading controls, respectively. (F) Protein model of the NifU N‐terminal domain and NifU C‐terminal domain of the NFU1 structure. Positions of amino acids affected by NFU1 missense variants are indicated in orange.
