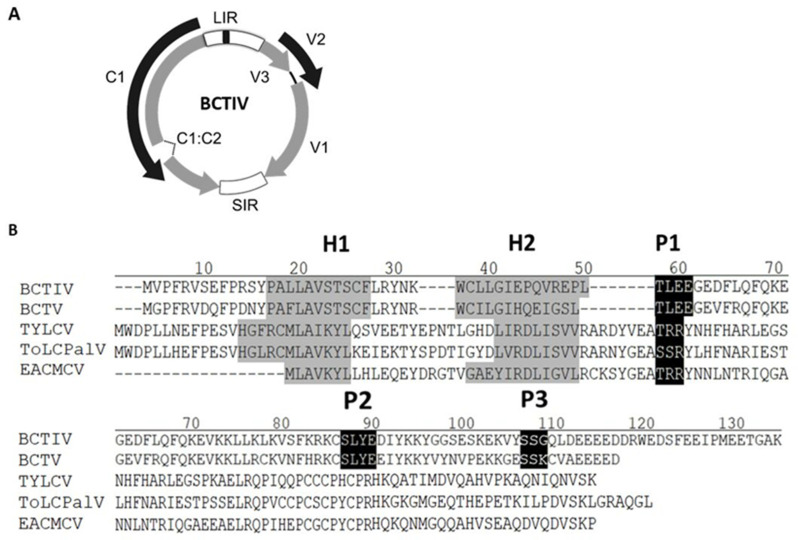
Figure 1.
(A): Beet curly top Iran virus (BCTIV) genome structure. Arrows represent open reading frames (ORFs). LIR: long intergenic region; SIR: short intergenic region. Common region inside LIR is depicted in black (B): Alignment of the aminoacid sequences of the V2 proteins from the becurtovirus beet curly top Iran virus (BCTIV; AFK14083), the curtovirus beet curly top virus (BCTV; AAA42752.1), the begomoviruses tomato yellow leaf curl virus (TYLCV; CAA33687.1), tomato leaf curl Palampur virus (ToLCPalV; CAP03292), and East African cassava mosaic Cameron virus (EACMCV; AF112354). Gaps (-) were introduced to optimize the alignment. The positions of the predicted putative phosphorylation motifs P1 (protein kinase CK2/protein kinase C), P2 (protein kinase CK2), and P3 (protein kinase C) are depicted in white letters inside black boxes. The hydrophobic domains (H1 and H2) are shadowed in gray.