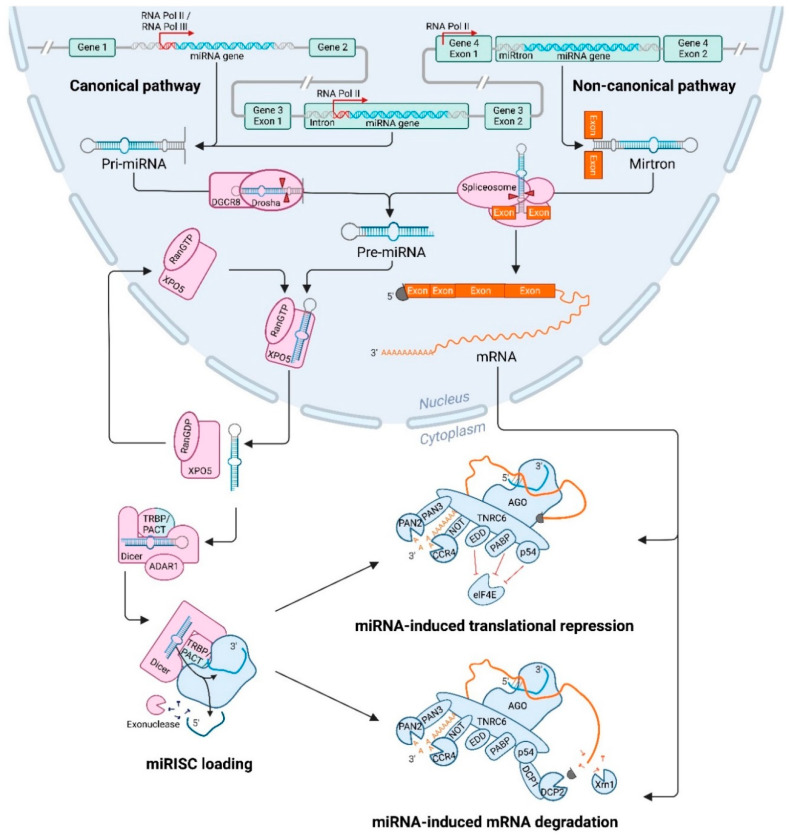
Figure 1.
General overview of the canonical and non-canonical route from microRNA gene to regulation of target messenger RNA. Following the canonical pathway of miRNA biogenesis, an intergenic (left), or intragenic (middle) miRNA gene is transcribed by RNA polymerase (Pol) II or III, to a primary-miRNA (pri-miRNA) transcript. This stem-loop transcript is cleaved by ribonuclease III enzyme Drosha and its co-factor DiGeorge Syndrome Critical Region 8 (DGCR8) to form a precursor-miRNA (pre-miRNA), which is subsequently exported out of the nucleus by Exportin-5 (XPO5) and its co-factor Ran-GTP. The non-canonical pathway of miRNA biogenesis involves the splicing of a pre-miRNA molecule from an entire intron without the need for Drosha processing, after which it enters the canonical pathway. In the cytoplasm, the pre-miRNA is cleaved by ribonuclease III enzyme Dicer and its co-factors transactivation-response element RNA binding protein (TRBP), protein kinase RNA activator (PACT) and adenosine deaminases acting on RNA 1 (ADAR1) to form a mature miRNA duplex. After strand selection, a single-stranded guide miRNA is incorporated into Argonaute (AGO), which is bound by trinucleotide repeat containing 6 protein (TNRC6) and then the functional miRNA-induced silencing complex (miRISC) is formed. The passenger strand is released and degraded by an exonuclease. After association with multiple co-factors and miRISC effector proteins, the miRISC is able to silence its target. The miRNA is hereby used as a guide, and based on complementarity between miRNA and messenger RNA (mRNA), translation is repressed or the mRNA is degraded. Binding of the miRISC to the target mRNA can induces deadenylation by CCR4-NOT as main effector, which is in some cases followed by decapping by DCP1/DCP2 and degradation by exonuclease XRN1 (Created with BioRender.com).