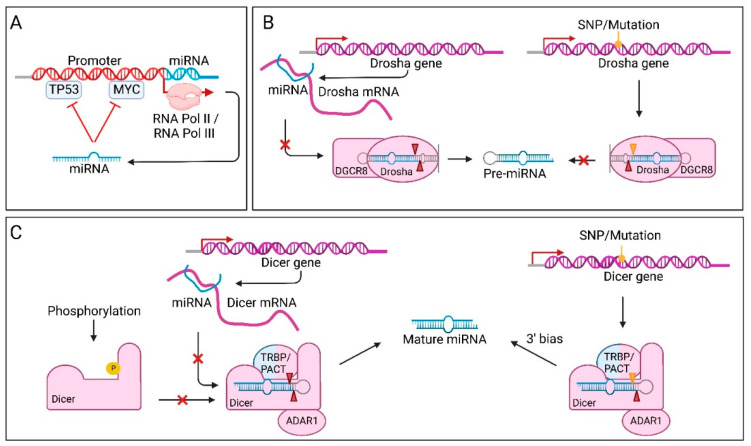
Figure 3.
The effect of altered transcription and altered processing through ribonuclease III enzymes Drosha and Dicer malfunctioning. This figure shows how steps of the biogenesis affect production of microRNA (miRNA). (A) Often, miRNAs are transcribed by RNA Polymerase II, which requires transcription factors, such as TP53 and MYC. These transcription factors can be targets of the transcribed miRNA, resulting in a feedback loop. (B) The Drosha RNase III enzyme processes primary miRNAs to precursor miRNAs (pre-miRNA). The protein’s functionality can be altered by many factors, such as single nucleotide polymorphisms (SNPs) and mutations. Drosha messenger RNA can also be the target of specific miRNAs, leading to a negative feedback loop in miRNA maturation. (C) The Dicer RNase III enzyme is able to process pre-miRNA to form a mature miRNA duplex. SNPs and mutations in the Dicer gene can alter its function, leading to altered pre-miRNA processing. Mutations in the RIIID domain of Dicer lead to problems with 5′ strand processing, inducing a 3′ strand bias. Besides inhibition, some SNPs are also capable of increasing Dicer effectivity. Dicer can also be regulated by specific miRNAs, leading to lower miRNA maturation. Phosphorylation of Dicer can induce nuclear translocation, leading to altered miRNA processing in the cytosol (Created with BioRender.com).