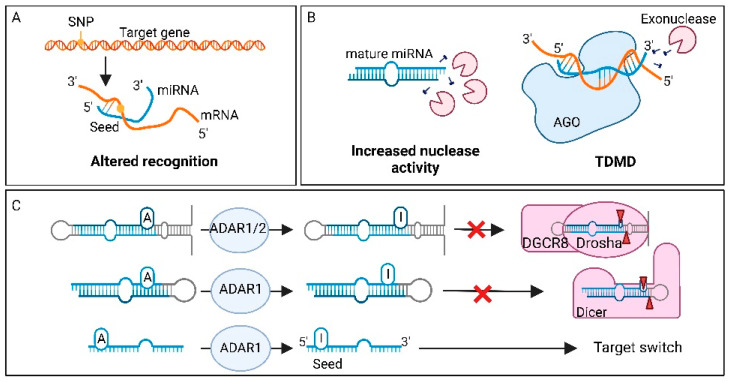
Figure 5.
microRNA dysregulation in cancer through target gene modifications, and factors affecting microRNA degradation and microRNA editing. (A) A mutation in the microRNA (miRNA) recognition site of a target gene can result in multiple changes; the target can become undetectable for the miRNA, the mutation might change the degree of repression or make a target recognizable for miRNA. (B) miRNA stability in the cell is important for regulation of miRNA targets, and stability is lowered by increased nuclease activity or target-directed miRNA degradation (TDMD). The latter process, TDMD, revolves around complementarity between target and miRNA. When a miRNA binds to its target with high 3′ end complementarity, the conformation of Argonaute (AGO) may change and the miRNA may become susceptible to degradation by exonucleases. (C) RNA editing, such as adenosine (A) to inosine (I) editing by adenosine deaminases acting on RNA 1/2 (ADAR1/2), can change the sequence of the RNA, with diverse consequences. miRNA editing can alter primary-miRNA and pre-miRNA in such a way, that they become unavailable for processing by ribonuclease III enzymes Drosha and Dicer, respectively. Additionally, the seed can also be edited, which allows for a shift in targetome, or altered recognition of existing targets (Created with BioRender.com).