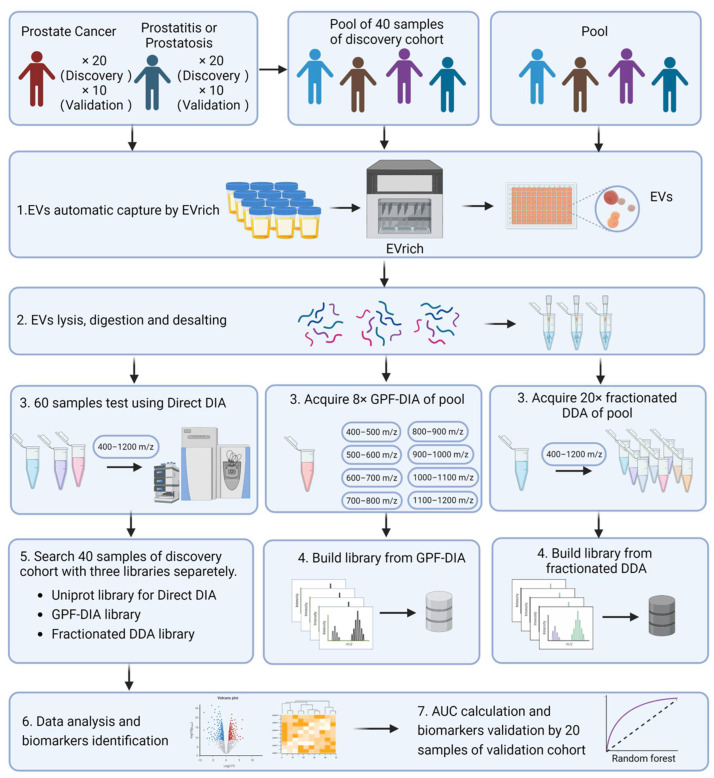
Figure 1.
Experimental Design and Workflow of the Study. (Created by BioRender.com) EVs were separated in urine samples collected from all participants. We then used the EVTRAP method to capture and isolate the EVs. For the establishment of the library for MS searching, in this step, the samples from all 40 participants of the discovery cohort were pooled as the main library for GPF-DIA-base search, while other independent 40 biological samples were pooled and conducted as a secondary library for fractionated DDA-base DIA search. Meanwhile, the particle number and size of EVs were examined using Nanoparticle tracking analysis (NTA). In addition, the size and structure of extracted EV were examined by morphology using Transmission electron microscopy (TEM). Subsequently, the isolated EVs were lysed, digested, and desalted into peptides, then three strategies were performed for MS measurement and searching separately. The individual EV proteins were analyzed for discrimination between prostate cancer cases and non-prostate cancer controls, with the annotation on potential mechanistic pathways. Lastly, a random forest model for accessing the performance of classifying prostate outcomes was performed.