Figure 1.
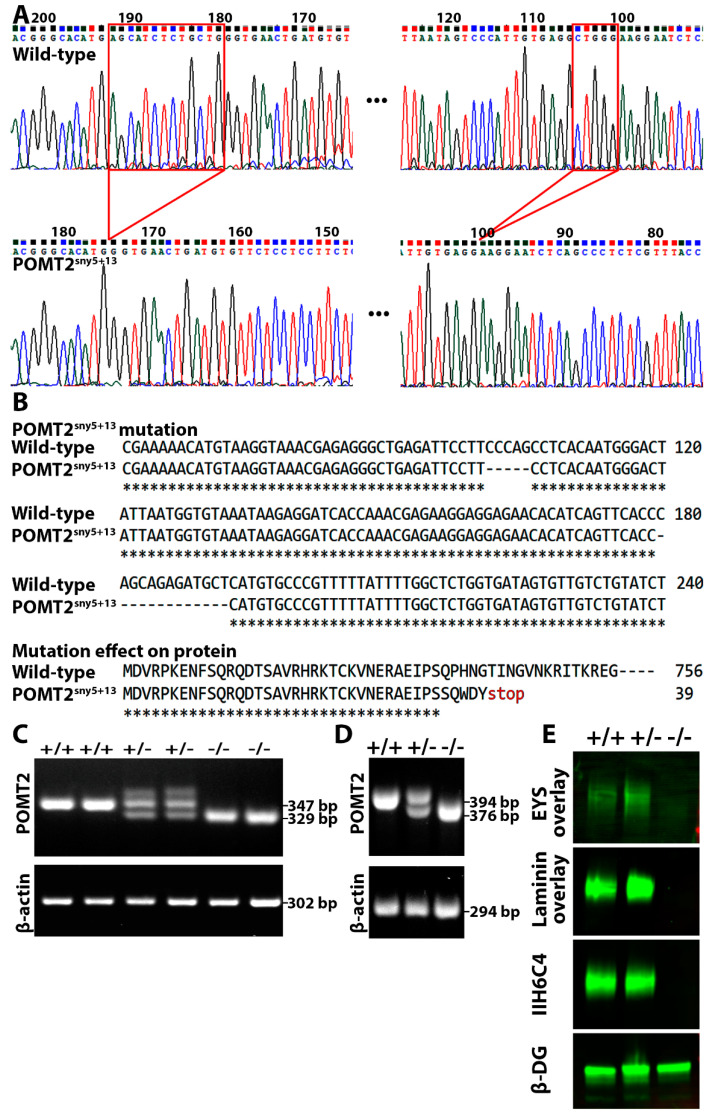
Deletion of pomt2 in zebrafish ablated matriglycan and caused loss of EYS binding. pomt2 mutant zebrafish were generated by CRISPR. Crossing of heterozygous mutant zebrafish generated homozygous mutant fish. Skeletal muscle lysates were used for IIH6C4 immunoblotting and EYS overlay assays. (A) Sequencing chromatogram of wild-type and pomt2sny5+13 PCR product. The two small deletions in exon 2 are bracketed with red marks. (B) Alignment of part of exon 2 sequences showing the two small deletions in the pomt2sny5+13 mutant and alignment of wild-type POMT2 protein with the expected truncated POMT2 peptides in the pomt2sny5+13 mutant. The mutant POMT2 protein has only 39 amino acid residues in total. (C) Example of PCR genotyping showing that wild-type zebrafish produce a 347 bp fragment while homozygous mutant animals produce a 329 bp fragment. (D) RT-PCR of wild-type animals produce only a 394 bp fragment, homozygous mutant animals produce a 376 bp fragment, and heterozygous animals produce both 394 and 376 bp fragments. (E) IIH6C4 and β-DG Western blotting and EYS and laminin overlay on pomt2sny5+13 mutant. Note abolished IIH6C4 reactivity and EYS and laminin binding in muscle lysate of homozygous pomt2 mutant.