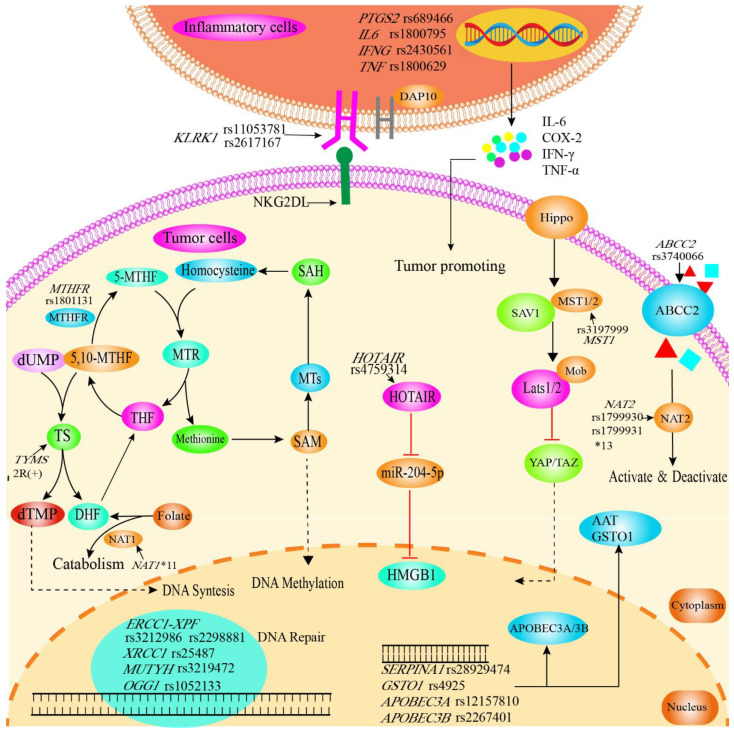
Figure 4.
Graphical synopsis of the role of SNPs in susceptibility to cholangiocarcinoma. SNPs in genes related to inflammation, DNA repair, cellular protection against toxins, RNA, enzymes and membrane proteins are related to susceptibility. Polymorphisms in inflammatory genes (IL6 rs1800795, INF-γ rs2430561, TNF-α rs1800629) affect levels of inflammatory cytokines, which play a key role in the inflammation-to-tumor process. The KLRK1/NKG2D receptor plays an important role in the surveillance of tumors by NK cells, and is induced by tumorigenic actions and further upregulated by chemotherapy or radiation. Polymorphisms in KLRK1 (rs11053781, rs2617167) might therefore facilitate immune escape and tumor progression. MTHFR plays an important role in folate metabolism and is also key in maintaining the balance between DNA synthesis and methylation. MTHFR rs1801131 reduces the activity of MTHFR, thereby leading to folic acid deficiency and can subsequently be linked to cancer risk. The Hippo signaling pathway mainly regulates cell proliferation and apoptotic and can be affected in its activity by MST1 rs3197999. ABCC2 is a member of the MRPs subfamily of the ABC transporter protein superfamily, transporting various molecules through extracellular and intracellular membranes. Therefore, reduced production of the ABCC2 protein by ABCC2 rs3740066 polymorphism affects the absorption, distribution and excretion of the substrate drug or toxic substance. SNPs in NAT1 and NAT2 can lead to reduced expression, decreased activity and/or instability of enzymes associated with the metabolism of drugs and carcinogenic substances. HOTAIR/miR-204-5p/HMGB1 regulates cell proliferation, apoptosis and autophagy, with SNPs potentially affecting regulation. Further, SNPs in DNA repair genes can naturally be associated with cancer susceptibility (ERCC1 rs3212986, rs2298881; MUTYH rs3219476; XRCC1 rs3219472; OGG1 rs1052133). Susceptibility to cancer is further influenced by SNPs in enzymes (APOBEC3A rs12157810, DNA editing; APOBEC3B rs2267401, RNA editing; SERPINA1 rs28929474, abnormal expression of AAT which acts as endogenous protease inhibitor; GSTO1 rs4925, maintenance of cellular redox homeostasis). AAT, α1-antitrypsin; ABCC2, ATP-binding cassette sub-family C member 2; APOBEC3A, Apolipoprotein B mRNA editing enzyme catalytic subunit 3A/3B; DAP10, DNAX activating protein 10; DHF, dihydrofolate acid; dTMP, deoxythymidine monophosphate; dUMP, deoxyuridine monophosphate; ERCC1, ERCC excision repair 1; GSTO1, Glutathione S-transferase omega 1; HMGB1, high mobility group box 1; HOTAIR, lncRNA HOX transcript antisense RNA; IFNG/IFN-γ, Interferon gamma; IL6/IL-6, Interleukin 6;KLRK1, Killer cell lectin-like receptor K1; LATS, large tumor suppressor kinase; miR-204-5p, microRNA-204-5p; MST1, Macrophage stimulating 1; 5, 10-MTHF, 5, 10-methylene THF; 5-MTHF, 5-methyl THF, MTHFR, methylenetetrahydrofolate reductase; MTs, methyltransferases; MTR, methionine synthase; Mob, Maps one binder; NAT1/2, N-acetyltransferase 1/2; NKG2D-L, NKG2D ligands; OGG1, 8-oxoguanine DNA glycosylase; PTGS2, Prostaglandin–endoperoxide synthase 2; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; SAV1, Salvador 1; TAZ, transcriptional coactivator with PDZ-binding motif; THF, tetrahydrofolate acid; SERPINA1, Serpin family A member 1; TNF-α, Tumor Necrosis Factor alpha; TS, thymidylate synthetase; XRCC1, X-ray cross complementation protein 1; YAP, Yes-associated protein.