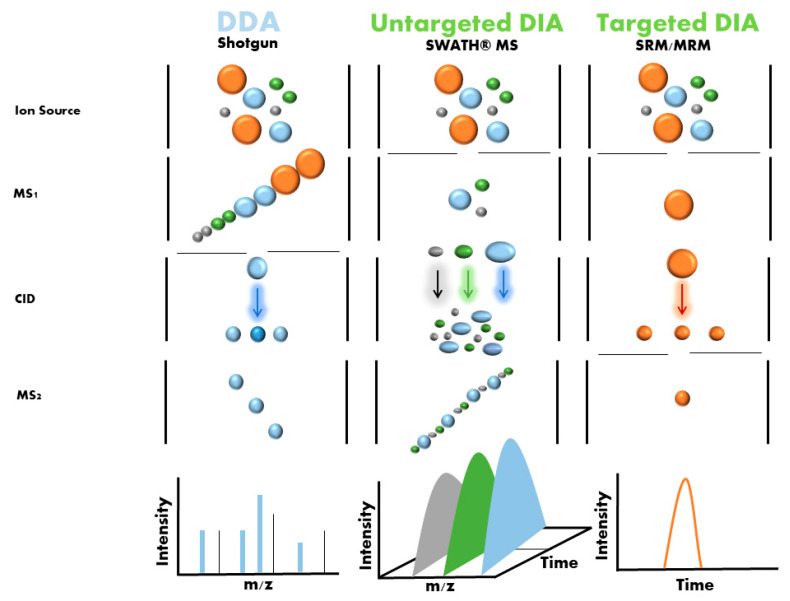
Figure 4.
DDA and DIA mode in proteomic analysis. In DDA mode, extracted proteins are digested and directly analyzed by single-shot DDA. The acquired data is searched against a database of known protein sequences and further processed using diverse software tools. In DIA mode, two distinct forms of proteomic analysis can be performed: SWATH-MS (untargeted DIA) and SRM/MRM (targeted DIA). In SWATH-MS extracted proteins are digested and directly analyzed by single-shot analysis in order to generate the spectral library. Once the library is generated, a multi-window running method is applied to individual samples; wide isolation windows span the entire MS1 m/z range and all precursor ions in the library that are found in each isolation window are fragmented in MS2. All proteins found in the library are quantified (untargeted) and all samples are processed using data analysis software to obtain the quantitative data. In MRM and SRM, the precursor ions to be analyzed are fixed (targeted) by the user, and only these are fragmented in MS2.