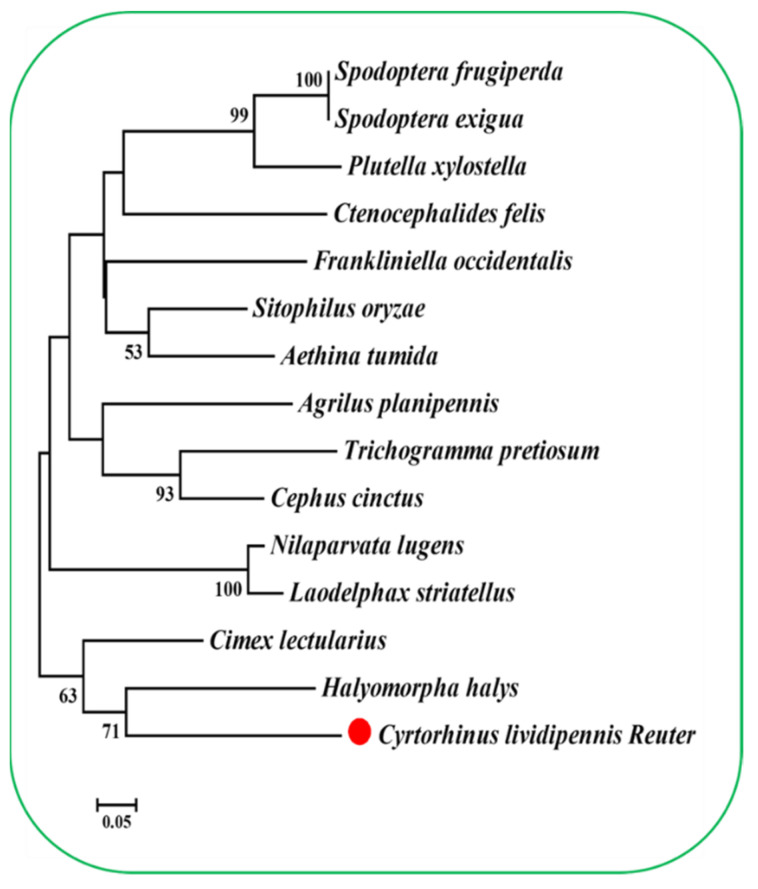
Figure 2.
The phylogenic analysis of phosphoserine phosphatases (PSPs). An unrooted phylogenetic tree is constructed by the maximum-likelihood method based on the protein sequence alignments. The PSP-like sequences originate from hemipteran species Cyrtorhinus lividipennis, Cimex lectularius (XP_024081153), Halyomorpha halys (XP_014271743), Nilaparvata lugens (AGG09860), and Laodelphax striatellus (AGC92248); coleopteran species Agrilus planipennis (XP_01833970), Aethina tumida (XP_019877426), and Sitophilus oryzae (XP_030747197); lepidopteran species Plutella xylostella (NP_001296070), Spodoptera frugiperda (XP_035439715), and Spodoptera exigua (KAF9423892); hymenopteran species Cephus cinctus (XP_024943432) and Trichogramma pretiosum (XP_014230438); siphonaptera species Ctenocephalides felis (XP_026462493); and thysanoptera species Frankliniella occidentalis (KYP96486.1). The bootstrap value (1000 replicates) for each node is shown. The scale bar corresponds to a distance of 0.05. The scale bar represents the amino acid divergence. The red point represents C. lividipennis Reuter PSP.