Figure 5.
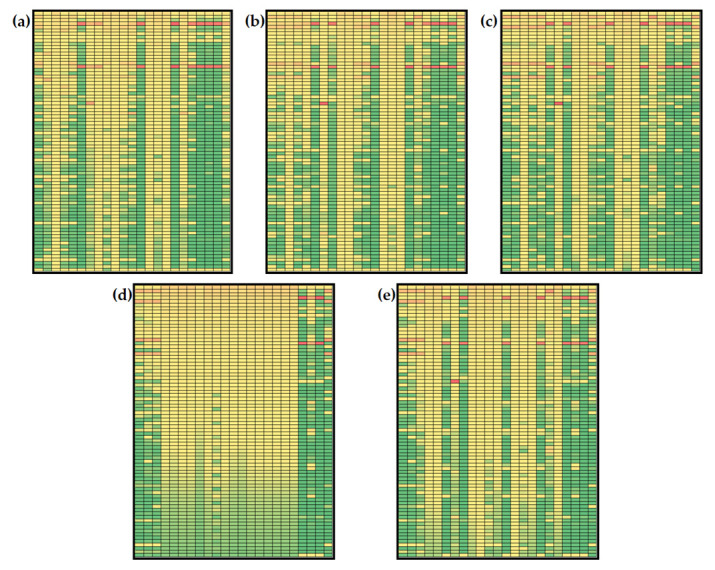
Global sensitivities of hepatic lipid model metabolites (initial conditions for simulations according to Menu 1): (a) tf = 10 min, (b) tf = 50 min, (c) tf = 100 min, (d) tf = 250 min, (e) tf = 500 min. Rows: βG, β6, βf, βl, βm, µAMP, µe, µs, µ1, µ2, µ3, µ4,c0, cc, dBA, k10, k11, k12, k13, k14, k22, k5, k6, k6l, k6p, k7, k8, k9, k9a, ka, kaa, kai, kal, kba, kbl, kbm, kcl, kcm, kd, kdl, kdy, kft, kga, kgi, kgl, kgl2, kgm, kgm2, kgp, klp, kLG, kLH, kMH, kna, kp, kp6, kpp, kr, kre, kt, kyl, kym, lmax, mmax, vmin, v10, v12, v6, v8, v9, vLG, vLH, vMH, y0, αG, αF, ms, me. Columns: I, GL, YL, PL, RL, AL, SL, TL, GM, YM, PM, RM, AM, TM, P, TA, AA, LA, GA, TCB, ANB, TLB, GB. The global sensitivity coefficients are marked with colors: green indicating low values of sensitivity coefficients, and red indicating high values of global sensitivity coefficients based on the data distribution at the percentile curve.
