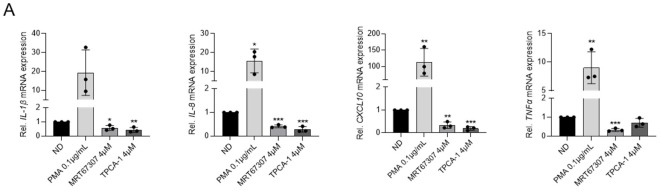
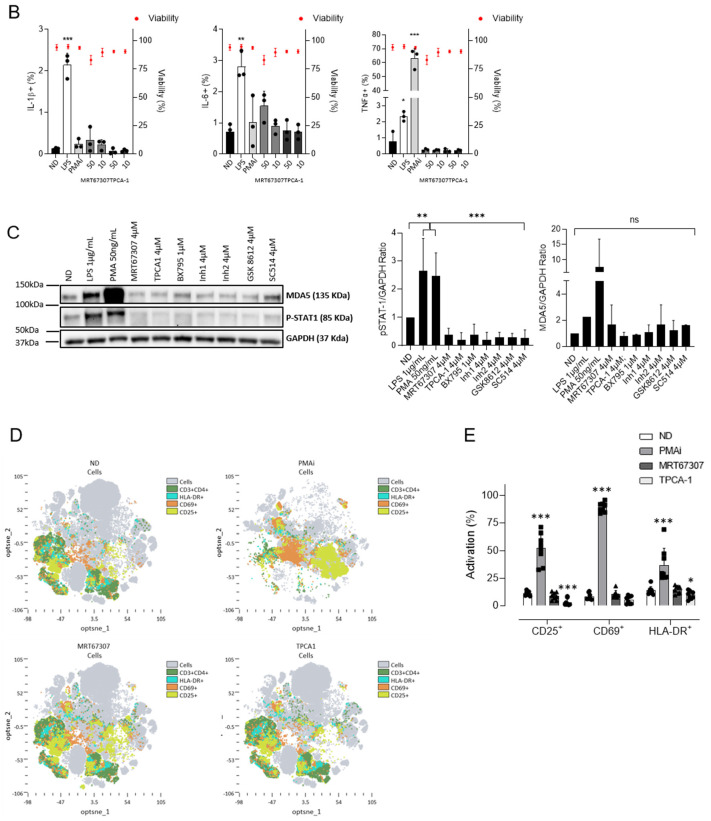
Figure 2.
Treatment with IKKi blocks production of inflammatory signals and T-cell activation. (A) Gene expression of proinflammatory cytokines IL-1β, IL-8, CXCL10, and TNFα in HL-60 cells after 16 h of treatment, as measured by RT-qPCR. PMA (0.1 µg/mL) was used as positive control. (B) Expression of proinflammatory cytokines IL-1β, IL-6, and TNFα as measured by intracellular staining by flow cytometry after 24 h of treatment in CD4+ T cells from HIV-negative donors. Viability of CD4+ T cells treated with IKKis as measured by flow cytometry. Black dots represent individual data from independent experiments. Mean ± SD of three independent experiments is shown. (C) Evaluation of innate immune activation at protein level. Representative western blot (out of three) showing the inhibition of distinct ISG upon treatment with IKKi (left panel). Semiquantitative analysis of ISG expression upon treatment with IKKi (right panel). (D) Distribution of cell surface markers in CD4+ T cells from uninfected donors treated with subtoxic concentrations of MRT67307 (4 µM) and TPCA-1 (4 µM) measured by flow cytometry. Distribution was determined by Opt-tSNE-guided manual gating analysis of the signal strength of key phenotypic defining cell activation markers CD25, CD69, and HLA-DR from eight HIV-negative donor samples. Opt-tSNE analysis was performed using 1000 iterations, a perplexity of 30, a trade-off θ of 0.5 as implemented by OMIQ data analysis software (www.omiq.ai). (E) Quantification of the cell activation markers compared to resting and PMAi activated CD4+ T cells. Each symbol refers to a distinct patient. Mean ± SD of n = 8 individuals is shown. * p < 0.05; ** p < 0.01; *** p < 0.001.