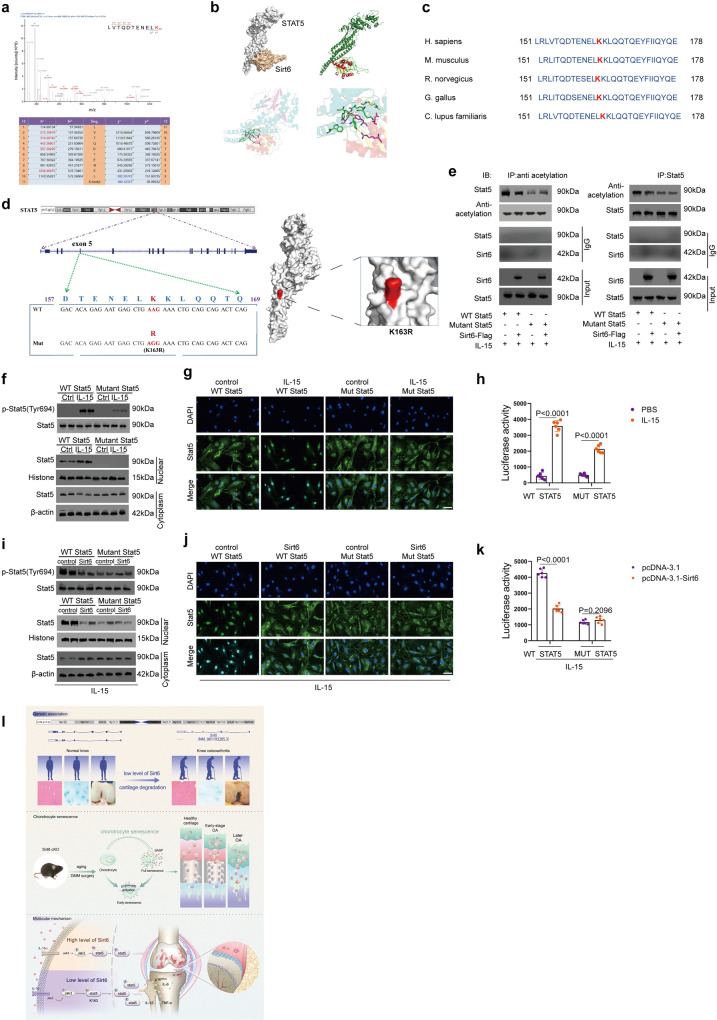
Fig. 5. Sirt6 modulates IL-15/JAK3/STAT5 signaling pathway by deacetylating lysine 163 (K163) on Stat5.
a LC-MS/MS analysis identified Sirt6 deacetylated K163 on Stat5. Lysates from human chondrocytes with or without Sirt6 treatment. b Interaction model between Stat5 and Sirt6. c Conservation of Stat5 K163 in different species. d K163 is located in exon 5 of STAT5, which was mutated to arginine through “A” replaced by “G”. The surface from molecular simulation showed the mutant site (K163R). e Sirt6 deacetylated Stat5 via K163. The acetylation level of WT Stat5 with Sirt6 treatment was analyzed by western blot. n = 3 independent biological replicates per group. f, g Human chondrocytes were transfected with WT Stat5 or mutant Stat5, followed by IL-15 treatment. The phosphorylation of Stat5 and the cellular localization of Stat5 was determined by western blot. n = 3 independent biological replicates per group (f). Immunofluorescent staining of Stat5. n = 3 independent biological replicates per group (g). h Transcriptional activity of WT and mutant Stat5. Human chondrocytes were co-transfected with WT Stat5 or mutant Stat5, followed by IL-15 treatment. n = 6 independent biological replicates per group. i, j K163 of Stat5 is required for Sirt6-regulated IL-15/JAK3/STAT5 signaling pathway. After IL-15 treatment, the phosphorylation of Stat5 and the cellular localization of Stat5 were analyzed. n = 3 independent biological replicates per group (i). Immunofluorescent staining of Stat5. n = 3 independent biological replicates per group (j). k The transcriptional activity of WT and mutant Stat5 in human chondrocytes transfected by pcDNA3.1 or pcDNA3.1-Sirt6, followed by IL-15 treatment. n = 6 independent biological replicates per group. l Molecular model for dysregulated Sirt6 in chondrocyte senescence and OA pathogenesis. Scar bar: g, j 20 μm. Data were presented as the mean ± s.e.m. P values are from two-tailed unpaired Student’s t-test (h, k). Source data are provided as a Source Data file.