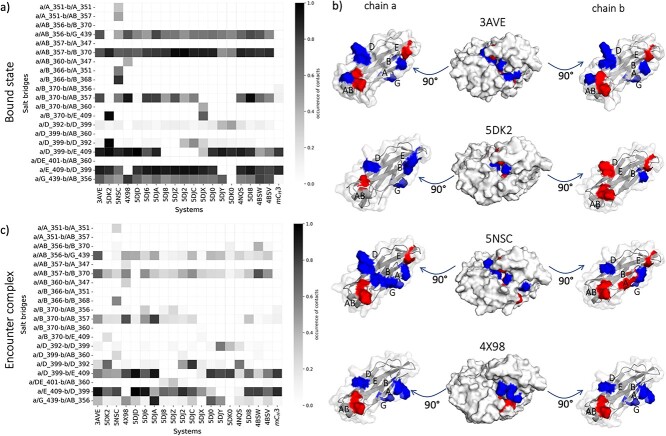
Fig. 3.
Comparison of salt bridges in the bound structures of the systems. a) Salt bridge occurrence between chain a and chain b of all the systems during the metadynamics simulations, until the start of the dissociation. The residues involved in a salt bridge are listed on the vertical axis of the matrix. The residue number is accompanied by a first letter (a or b), representing the chain, and a capital letter, representing the strand or loop in which the residue is located. b) The pictures show the open interface of the wildtype and of systems with charged mutations (PDB codes, respectively: 3AVE, 5DK2, 5NSC, 4X98). The residues in the interface that make salt bridges are colored: in blue the residues with a positive charge and in red the ones with a negative charge. c) Salt bridge occurrence between chain a and chain b when the systems are in the encounter complex states, colored according to their occurrence.