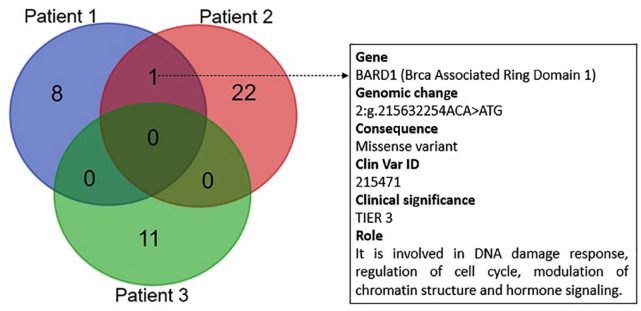
Figure 2.
Venn diagram reporting the number of shared genetic variant between the analysed patients. The most common genetic variant was BARD1 p.Val507Met. Gene nomenclature, genomic change, consequence, Clin Var ID, priority and biological role are reported to the right of the diagram. DNA was extracted form FFPE primary tissue specimens through the MGF03-Genomic DNA FFPE One-Step Kit (MagCoreDiatech). Libraries were prepared with TruSigtTMOncology 500 kit that analyses 523 cancer-relevant genes and sequenced on an Illumina NovaSeq 6000 (San Diego, USA) platform. A median of 90 million reads were generated for each sample and the coverage in the target region was above manufacturer’s suggested threshold of 150×. Sequences were aligned to the human reference genome GRCh37 (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/index.shtml) using the Burrows–Wheeler Aligner tool. The prioritization of variants was done according to a four-tiered structure, adopting the joint consensus recommendation by AMP/ACMG (Association for Molecular Pathology and American College of Medical Genetics).
AMP, Association for Molecular Pathology; FFPE, formalin-fixed and paraffin-embedded.