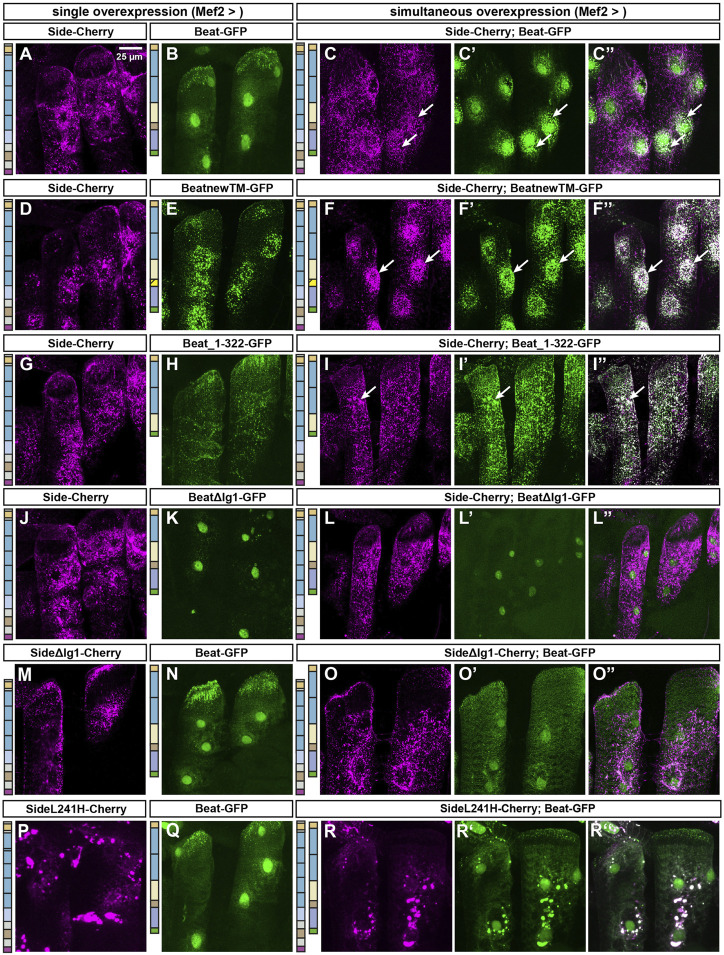
FIGURE 5.
The first immunoglobulin domains of Beat and Side mediate their mutual interaction. (A–Q) Confocal images showing lateral muscles 21 and 22 in third instar larvae imaged through the translucent cuticle of intact third instar larvae using a ×63 oil objective. The two columns to the left show the distribution of either Side-Cherry (magenta) or a given Beat fusion protein (GFP, green) expressed individually in muscles. The three columns to the right show simultaneous co-overexpression of the indicated combination of two constructs. (A–C'') Side-Cherry forms punctate aggregates but reaches also the muscle surface (A). Beat-GFP mostly accumulates in muscle nuclei (B). Simultaneous co-overexpression of both proteins re-distributes Beat-GFP to cytoplasmic aggregates surrounding nuclei, where portions of both proteins co-localize (arrows in C-C''). (D–F'') BeatnewTM-GFP completely traps Side-Cherry in intracellular particles surrounding nuclei (arrows). (G–I'') Beat_1–322-GFP co-localizes with Side-Cherry in punctate clusters throughout the sarcoplasm (arrows). (J–L'') BeatΔIg1-GFP accumulates in muscle nuclei, irrespective of whether or not Side-Cherry is co-expressed. (M–O'') SideΔIg1-Cherry shows a similar subcellular distribution as full-length Side (M). Co-expression with Beat-GFP results only in a few Beat-GFP punctae outside nuclei (O–O''). (P–R'') Side carrying the L241H point mutation clusters in large sarcoplasmic aggregates that do not reach the muscle surface (P). In co-overexpression experiments, Beat-GFP accumulates in these aggregates but enriches also in nuclei. Schemes to the left of each column indicate the domain structure of the constructs used for each experiment.Scale bar: 25 μm.