Figure 2. Identification of CPL1 as a fungal effector by forward genetics.
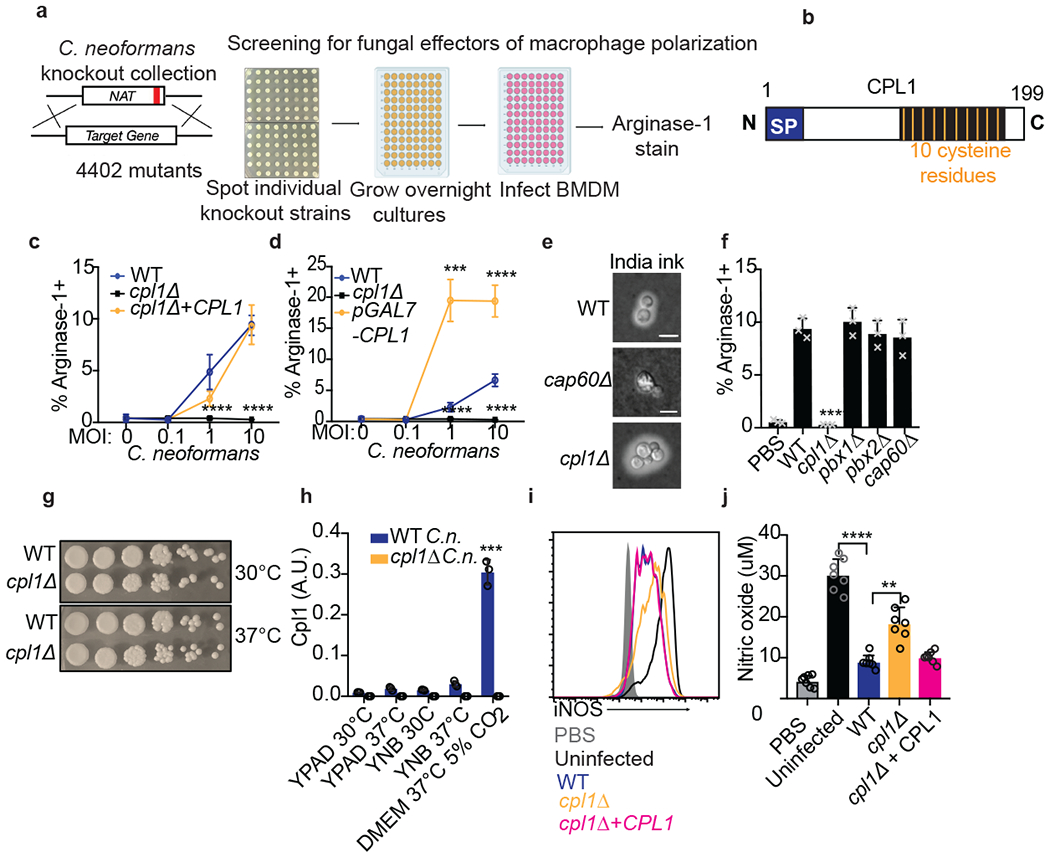
(a) Genetic screening strategy to identify fungal effectors that drive arginase-1 expression. (b) Outline of CPL1 protein domain architecture. (c) Complementation assay using arginase-1 FACS on BMDMs stimulated for 24hrs with either WT, cpl1Δ, or cpl1Δ + CPL1 C.n. strains at the indicated MOIs. n = four biologically independent samples. (d) Arginase-1 FACS on BMDMs stimulated for 24hrs with WT, cpl1Δ, or pGAL7-CPL1 C.n. strains at the indicated MOIs. n = four biologically independent samples. (e) India ink staining for capsular polysaccharides in the indicated strains after overnight culture in 10% Sabouraud media. Scale bar indicates 5 microns. Data are representative of two independent experiments. (f) Arginase-1 FACS on BMDMs stimulated for 24hrs with the indicated capsule mutant strains at MOI=10. n = three biologically independent samples. (g) Spotting assay for WT vs cpl1Δ C.n. growth on YPAD plates incubated at the indicated temperatures. (h) RT-qPCR for CPL1 mRNA expression in cultures grown to OD600=1.0 in the indicated conditions (A.U. = arbitrary units relative to ACT1). n = three biologically independent samples. (i) Representative FACS histogram for intracellular iNOS protein levels in BMDMs pre-infected with the indicated strains at an MOI=10 for 2hrs followed by 24hr stimulation with LPS (100ng/ml) and IFNγ (50ng/ml). (j) Total nitric oxide in supernatants from BMDMs treated as in (i). n = seven biologically independent samples. Data are presented as mean values +/− SD. **p < 0.01; ***p < 0.001; ****p < 0.0001 by one-way ANOVA with Bonferroni test.
