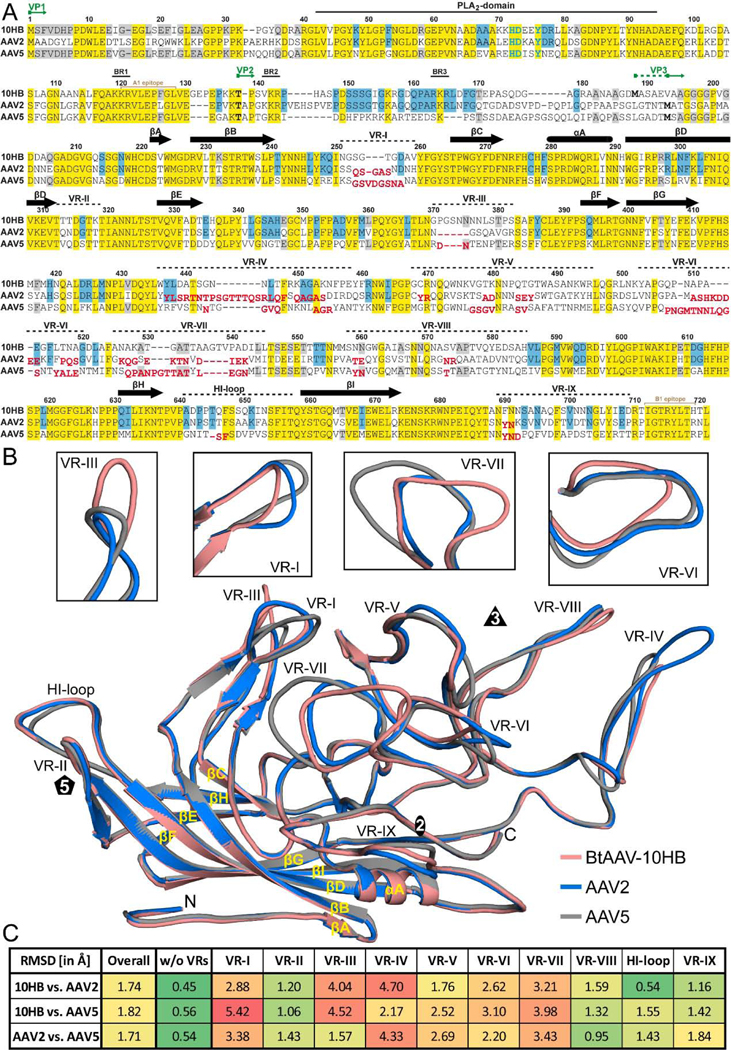
FIG. 6.
Capsid structure comparison of 10HB, AAV2, and AAV5. A) Structure-based sequence alignment, except for aa 1 to 209 (10HB numbering) which is based exclusively on the amino acid sequence. The VP1, VP2, and VP3 N-termini residue numbers based on 10HB VP1, the phospholipase (PLA2) domain, the basic regions (BR), and the location of the variable regions (VRs) are indicated above the amino acid sequence. Secondary structure elements such as β-strands and α-helices, are shown as black arrows and black cylinders, respectively. Amino acids highlighted in yellow indicate sequence identity among all three viruses whereas amino acids in blue or gray indicate an identical residue in 10HB compared to AAV2 or AAV5, respectively. Amino acids in AAV2 or AAV5 whose Cα atoms are further than 2 Å apart when superposed onto 10HB are shown offset and in red below the aligned residues. The A1 and B1 epitope are indicated in light brown. B) Structural superposition of 10HB (salmon), AAV2 (blue), and AAV5 (gray) shown as ribbon diagrams with the position of VR-I to VR-IX, the HI-loop, β-strand A-I, α-helixA, the N- and C-terminus, and the icosahedral 2-, 3-, and 5-fold axis labeled. Close-ups of VR-I, VR-III, VR-VI, and VR-VII are shown above separately for clarity. This image was generated using PyMOL (DeLano, 2002). C) Calculated root mean square deviations (RMSDs) of the VPs of 10HB, AAV2, and AAV5 to each other for the overall structure and for specific VRs.