FIGURE 1.
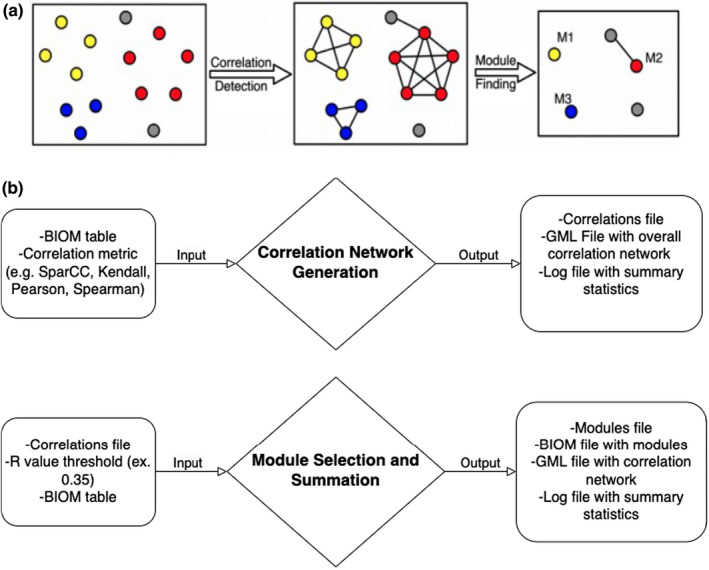
SCNIC schematic and data flow. (a) The basic process of SCNIC involves first identifying pairwise correlations between species and using them to build a correlation network. Modules of correlated features are identified and then summarized for downstream statistical analysis, or multi‐omic analysis between modules of microbes and other feature types. (b) The input to SCNIC comes in the form of a count table in BIOM format. The first step takes the table and generates a correlation table and network. The table is in a tab delimited format and the network is in GML format and can be used to visualize the network in Cytoscape. Modules are detected and summarized in the final step which generates a module membership file indicating which features are in each module. The collapsed BIOM table contains the same total counts per sample as the original table, but with less features. All features not included in modules are retained with their original counts and all modules have a total count per sample of the sum of the counts of all features in that module.
