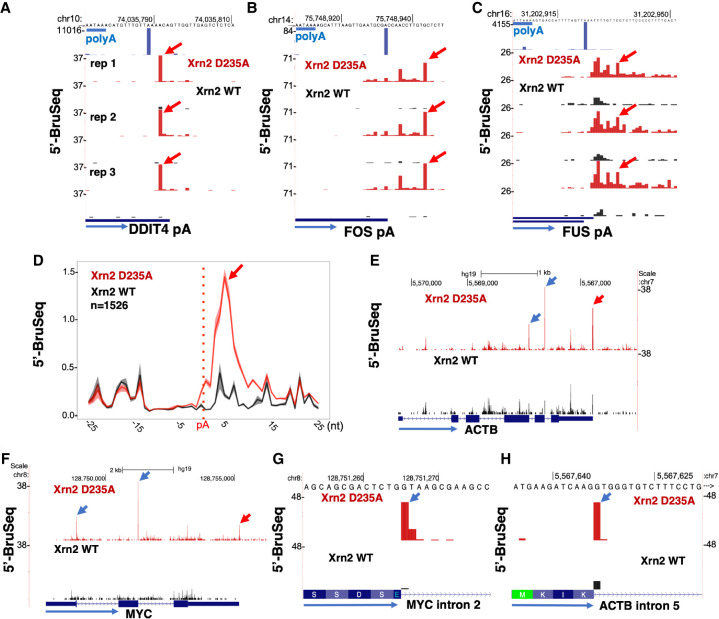
Figure 4.
Xrn2 loads several bases downstream from polyA sites. (A–C) Xrn2 D235A stabilizes 5′-PO4 ends (red arrows) relative to WT at the DDIT4 polyA site mapped by 3′ READS (top track, A) and several bases downstream from the FOS and FUS polyA sites (B,C). PolyA consensus is underlined in blue. Three replicate 5′-PO4 Bru-seq data sets are shown. (D) Metaplots of 5′-PO4 ends near polyA sites show Xrn2 engages a few bases downstream from the cleavage site. Reads were aligned to HEK293 polyA sites mapped by 3′ READS. Mean and range of three replicates. (E,F) Xrn2 inactivation stabilizes 5′-PO4 ends near 5′ splice sites (blue arrows) and polyA sites (red arrows). (G,H). Xrn2 degradation of debranched introns, suggested by stabilization of 5′-PO4 ends at the first base of the intron (blue arrow) in Xrn2 mutant cells.