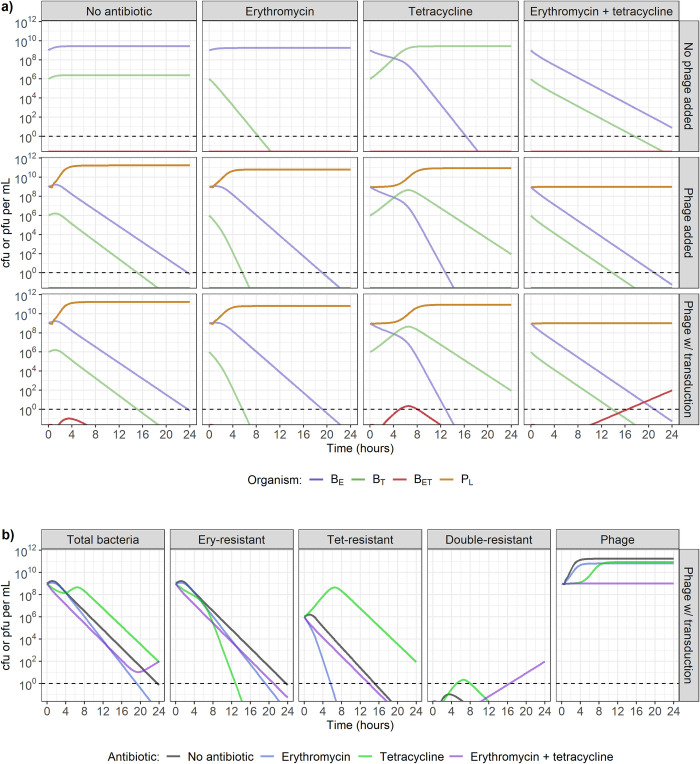
Fig 4.
a) Model-predicted dynamics with one single-resistant strains starting at carrying capacity (109 colony-forming units (cfu)/mL) and the second in minority (106 cfu/mL), in the presence of no antibiotics (1st column), erythromycin only (2nd column), tetracycline only (3rd column), or both erythromycin and tetracycline (4th column), combined with either no phage (top row), phage incapable of transduction (middle row), or phage capable of generalised transduction (bottom row). Erythromycin-resistant bacteria (BE) are initially present at a concentration of 109 cfu/mL, and tetracycline-resistant bacteria (BT) at 106 cfu/mL. Antibiotics and/or phage (PL) are present at the start of the simulation, at concentrations of 1 mg/L (4 x MIC for susceptible strains) and 109 plaque-forming units (pfu)/mL respectively. Double-resistant bacteria (BET) can initially be generated by generalised transduction only, and then by replication of existing BET. Dashed line indicates the detection threshold of 1 cfu or pfu/mL. b) Change in bacteria (single-resistant to erythromycin, single-resistant to tetracycline, or double-resistant) and phage numbers depending on the antibiotic exposure, in the presence of phage capable of generalised transduction. Dashed line indicates the detection threshold of 1 cfu or pfu/mL.