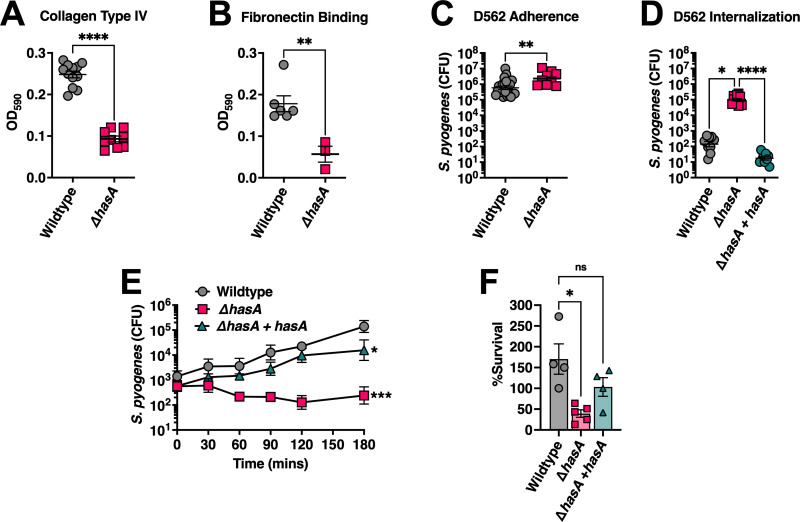
Fig 2. The S. pyogenes HA capsule inhibits host cell invasion but promotes survival from neutrophil-mediated killing.
Binding of S. pyogenes to wells pre-coated with 1 μg of human ECM components (A) fibronectin and (B) collagen type IV. (C) Adhesion of S. pyogenes to D562 pharyngeal epithelial cells. Confluent cell monolayers were cultured with S. pyogenes (MOI of 100) for 2 h at 37°C + 5% CO2. Cells were washed with PBS and lysed with Triton X-100 for enumerating remaining adherent bacteria. (D) Internalization of S. pyogenes into D562 cells. Confluent D562 cells were cultured with S. pyogenes (MOI of 100) for 2 h at 37°C + 5% CO2 followed by 1 h in media supplemented with 100 μg mL-1 of gentamycin. Bars represent mean CFUs ± SEM and each dot represents a biological replicate. Statistical differences were evaluated by unpaired t-test (A–C) (**, P < 0.01; ****, P < 0.0001) or (D) one-way ANOVA (*, P < 0.05; ****, P < 0.0001). (E) Whole human blood survival assay. Heparinized blood from human donors were inoculated with ~103 CFUs of S. pyogenes MGAS8232 at 37°C with rotation for 3 h. Data points represent geometric mean CFUs ± SD at each timepoint (n ≥ 3). Statistical significance was determined using one-way ANOVA with Friedman test (***, P < 0.001). (F) Neutrophil survival assay. Neutrophils were isolated from human blood by density centrifugation and inoculated with opsonized S. pyogenes at a MOI of 10. Surviving bacteria were enumerated after 60 mins at 37°C with rotation and calculated as the difference between survival in the no neutrophil control and in the presence of neutrophils. Each data point represents S. pyogenes CFUs from an individual donor. Data shown are the means of percent survival ± SD. Statistical analyses were performed using one-way ANOVA with Kruskal-Wallis test (*, P < 0.05).