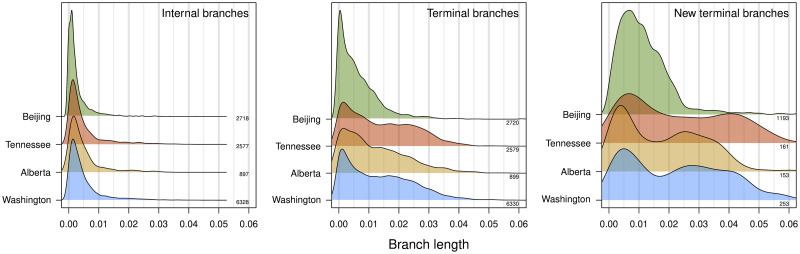
Fig 2. Distributions of branch lengths in HIV-1 pol phylogenies.
Each density summarizes the distribution of branch lengths (measured in units of expected nucleotide substitutions per site) for different locations and types of branches, as indicated in the upper right corner of each plot. We used Gaussian kernel densities with default bandwidths adjusted by factors 1.5, 0.75 and 0.75, respectively. Densities are labeled on the right with the corresponding number of branches. Internal and terminal branches are derived from the phylogeny reconstructed from background sequences. New terminal branches refer to additional branches to incident (new) sequences as placed onto the phylogeny by maximum likelihood (pplacer).