Figure 6.
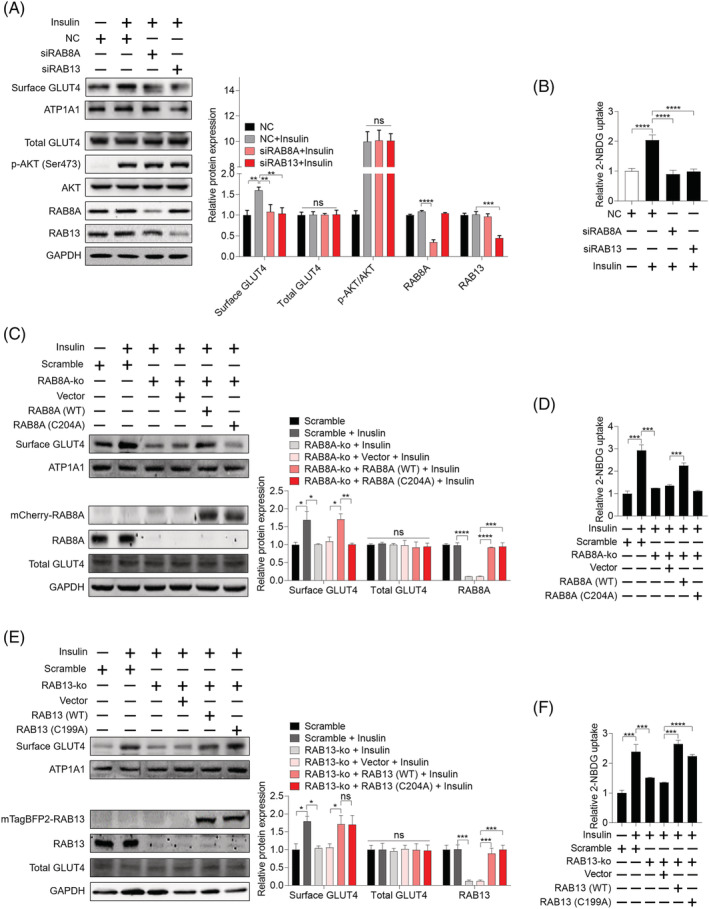
Geranylgeranylation of RAB8A is critical for insulin‐stimulated GLUT4 translocation and concomitant glucose uptake in skeletal muscle cells. (A) C2C12 myotubes were transfected with siRNAs specifically targeting RAB8A and RAB13, respectively, for 48 h. Then cells were incubated with 100 nM insulin for 30 min. Total protein samples and plasma membrane fraction samples were harvested, and the expression of indicated proteins was analysed by western blot, with GAPDH as the loading control (n = 3). (B) C2C12 myotubes were transfected with siRNAs specifically targeting RAB8A and RAB13, respectively, for 48 h. Then cells were exposed to 2‐NBDG containing 100 nM insulin for 30 min, and 2‐NBDG uptake was measured by fluorescence detection (n = 3). (C) RAB8A‐knockout (RAB8A‐ko) C2C12 myotubes were transfected with vector, RAB8A (WT), and RAB8A (C204A) plasmids for 48 h. Then cells were incubated with 100 nM insulin for 30 min. Total protein samples and plasma membrane fraction samples were harvested, and the expression of indicated proteins was analysed by western blot, with GAPDH as the loading control (n = 3). (D) RAB8A‐ko C2C12 myotubes were transfected with vector, RAB8A (WT), and RAB8A (C204A) plasmids for 48 h. Then cells were exposed to 2‐NBDG containing 100 nM insulin for 30 min, and 2‐NBDG uptake was measured by fluorescence detection (n = 3). (E) RAB13‐ko C2C12 myotubes were transfected with vector, RAB13 (WT), and RAB13 (C199A) plasmids for 48 h. Then cells were incubated with 100 nM insulin for 30 min. Total protein samples and plasma membrane fraction samples were harvested, and the expression of indicated proteins was analysed by western blot, with GAPDH as the loading control (n = 3). (F) RAB13‐ko C2C12 myotubes were transfected with vector, RAB13 (WT), and RAB13 (C199A) plasmids for 48 h. Then cells were exposed to 2‐NBDG containing 100 nM insulin for 30 min, and 2‐NBDG uptake was measured by fluorescence detection (n = 3). Data represented the mean ± SEM. Statistical analysis was performed with one‐way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns denotes no significance.
