Fig. 3. |. HG002 human trio polishing and evaluation.
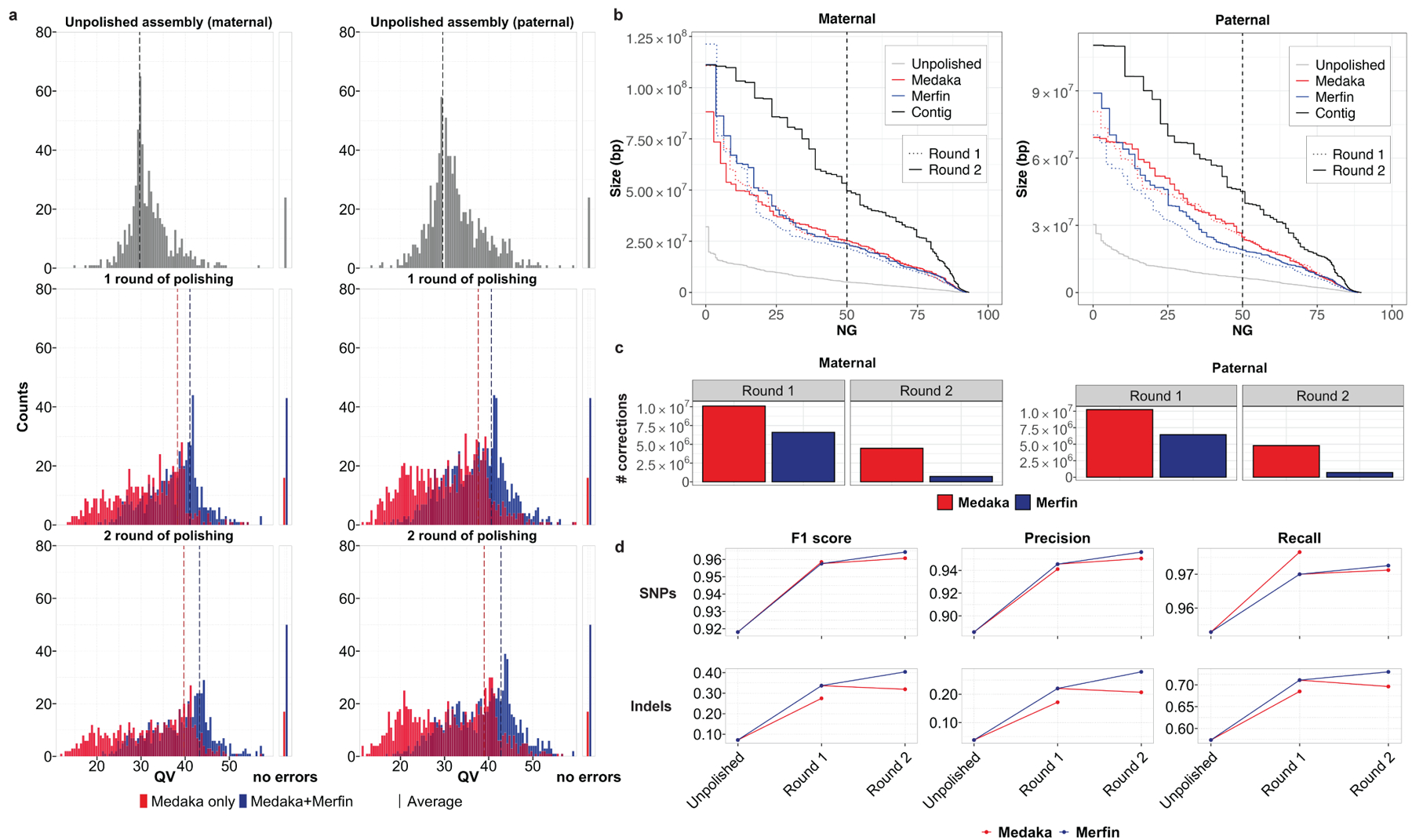
a, Distribution of QV scores as measured by Merqury for maternal and paternal contigs polished with Medaka only, or with variants generated by Medaka filtered with Merfin, from the experiment (test 4, Supplementary Table 3) using latest basecaller (Guppy 4.2.2) and highest coverage (~50x). The first panel represents the unpolished contigs, the mid panel the first round of Medaka polishing and filtering, and the last panel the second round applied to the Merfin results from the previous round. The number of contigs without evidence of errors as judged by Merqury QV are reported on the right side. b, Size of the haplotype blocks before and after polishing with or without Merfin for both the maternal and paternal assemblies. First round of polishing is represented by the dotted lines. c, Number of variants generated by Medaka for polishing and remaining variants after Merfin filtering for both the maternal and paternal assemblies. d, Assembly-based HG002 small variant calling performance of Merfin vs regular Medaka against GIAB truth set. Variants from the assembly are derived against GRCh38 using dipcall.
