To the Editor: Type 2 diabetes (T2D) and cholelithiasis are both common diseases with a high prevalence. In China, the prevalence of T2D is 0.9%[1] and that of adult cholelithiasis is 6.83%.[2] Obesity, insulin resistance, and abnormal metabolism are risk factors for T2D; epidemio-logic studies have suggested that they are also risk factors of cholelithiasis.[3,4] Several studies indicated that patients with diabetes are at an increased risk of developing cholelithiasis.[5] In recent years, increasing evidence has demonstrated an imbalance in the gut microbiota in both diabetes and cholelithiasis. In patients with T2D, the genera Bifidobacterium, Bacteroides, and Roseburia are decreased, whereas Ruminococcus, Fusobacterium, and Blautia are significantly increased. In patients with gallstone, intestinal bacteria (Bacteroides, Eubacterium, and Escherichia coli) involved in the oxidation and epimerization of bile acids can disrupt enterohepatic circulation and lead to cholelithiasis.[6,7]
Although the composition of the gut microbiota in patients with diabetes or cholelithiasis has been studied, the underlying role of the gut microbiota in the development from diabetes to cholelithiasis remains unclear. To this end, 16S ribosomal (RNA) gene amplicon sequencing was carried out to analyze the gut microbial composition of volunteers (Health group), T2D patients (Diabetes group), cholelithiasis patients (Stones group) and T2D complicated with cholelithiasis patients (Diastone group). Clarifying the gut microbial characteristics among different patient groups may improve the understanding of why patients with diabetes are more prone to developing cholelithiasis.
All patients with cholelithiasis were classified as having cholesterol gallstones.[8] Blood samples were collected on the first day of hospitalization, and total bile acids were tested using these blood samples. Genomic DNA was isolated from the feces and used to amplify the gene sequence in the V4 region of the bacterial 16S rRNA. Raw reads were obtained and subjected to quality analysis [Supplementary Figure S1]. The data reported in this paper have been deposited in the Genome Sequence Archive (CRA005196), accessible at https://ngdc.cncb.ac.cn/gsa.
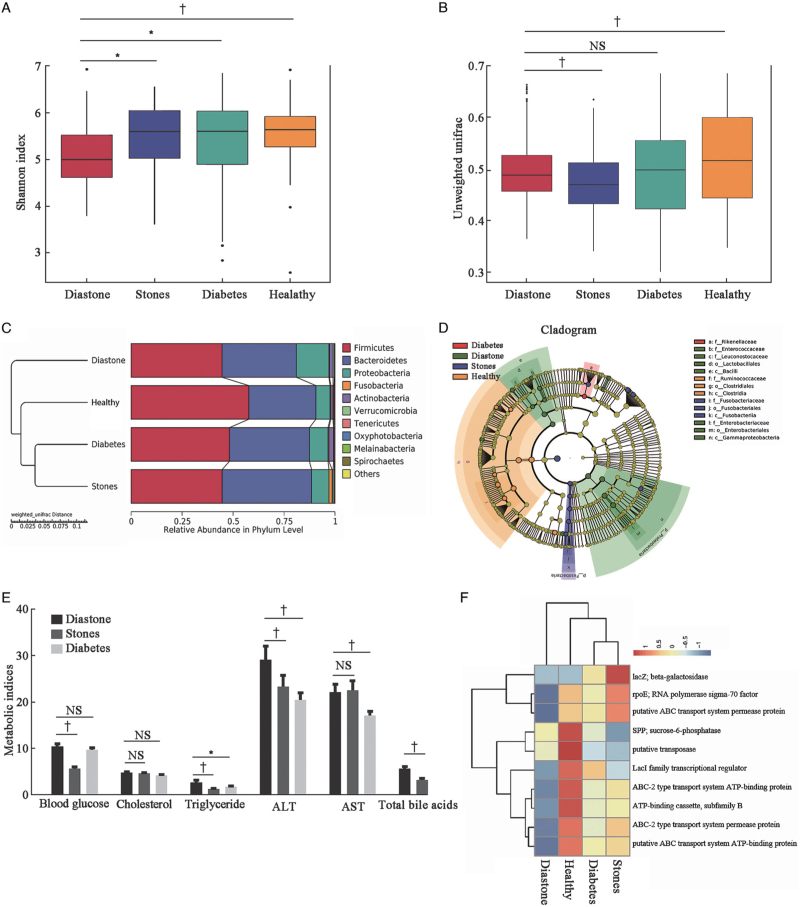
The intra-community alpha diversity, which represents the microbial richness, was analyzed. Among the four groups, the Shannon index of the diastone group was the lowest and significantly different from the other three groups (vs. stones group, P = 0.0224, vs. diabetes group, P = 0.0159, vs. healthy group, P = 0.0091, Wilcoxon rank-sum test) [Figure 1A]. Other alpha diversity indices were also analyzed [Supplementary Figure S2A and B]. Beta-diversity was evaluated to determine whether gut microbial community differences were significant between groups. The unweighted UniFrac-based beta diversity between the diastone and stone groups, diastone and healthy groups were significantly different (P < 0.01), whereas there was no significant difference between the diastone and diabetes groups (P = 0.3858, Wilcoxon rank-sum test) [Figure 1B]. The principal component analysis plot is shown in Supplementary Figure S2C. These results suggest that the diastone group share common gut microbial characteristics with the diabetes group.
Figure 1.
Characteristics of gut microbiota among four groups. (A) Intra-community alpha diversity with Shannon indices among four groups. (B) Unweighted unifrac based-beta diversity among four groups. (C) Weighted unifrac distance based-unweighted pair-group method with arithmetic mean (UPGMA) cluster tree analysis at the phylum level, left: UPGMA clustering tree structure, right: relative abundance distribution of species. (D) Different structures of gut microbiota among four groups by LEfSe. The abundance is proportional to the diameter of the circle. Linear discriminant analysis score was 3.5. (E) Typical metabolites in blood samples from different groups. (F) Heatmap of top ten function clusters by KO pathway (Kruskal-Wallis nonparametric test. The results are shown as the mean ± standard error of the mean. ∗P < 0.05 †P < 0.01). ALT: Alanine transaminase; AST: Aspartate transaminase; KO: Encyclopedia of Genes and Genomes orthology; LEfSe: Linear discriminant analysis effect size.
At the phylum level, the relative abundances of Firmicutes, Bacteroidetes, and Proteobacteria were >96% in all groups. Firmicutes was most common in the healthy group (approximately 57.75%), but decreased to 48.23%, 44.64%, and 44.60% in the diabetes, stone, and diastone groups, respectively. In contrast, Proteobacteria increased from 6.91% to 9.12% and 8.53% in the diabetes group and stones group, respectively, and even up to 15.95% in the diastone group. In the diastone group, the genus unidentified Enter-obacteriaceae was strikingly increased from 1.47% in the healthy group to 3.38%, 4.23%, and 5.40% in the diabetes, stones, and diastone groups, respectively [Figure 1C; detailed information in Supplementary Figure S3]. Linear discriminant analysis effect size (LEfSe) analysis was performed to detect biomarkers. Compared with the other three groups, the family Enterobacteriaceae, order Enterobacteriales, class Gammaproteobacteria, and phylum Proteobacteria in the diastone group were significantly increased [Figure S1D]. Interestingly, the specific bacteria showing significant differences in the diabetes group were similar to those in the diastone group [Supplementary Figure S2D]. Thus, the tendency of gut microbiota alterations in patients with T2D and cholelithiasis was similar but changes were more severe in patients with T2D and significantly different from those in patients with cholelithiasis.
Comparison of various metabolites in the blood samples of the subjects showed that although the diastone group had similar blood glucose levels as the diabetes group, they had higher triglyceride, alanine aminotransferase, aspartate aminotransferase, and total bile acid levels [Figure 1E; detailed information in Supplementary Table S1]. We performed correlation analysis of these clinical characteristics and altered gut microbiota and found that the phylum Tenericutes was positively with total bile acid and blood glucose levels. However, the genus Enterobacteriaceae was negatively with total cholesterol levels [Supplementary Table S2]. Using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) algorithm for functional analysis, Encyclopedia of Genes and Genomes orthology analysis showed that the genes for ribose nucleic acid (RNA) polymerase sigma-70 factor, ABC transport system permease protein, and transport system adenosine triphosphate (ATP)-binding protein were remarkably decreased in the diastone group compared to the other three groups [Figure 1F].
In our study, patients with T2D and cholelithiasis had a unique microbial community structure that was highly similar to that in patients with T2D. The genus unidentified Enterobacteriaceae and phylum Proteobacteria, which were remarkably increased in the diastone group, form a typical cluster of gram-negative facultative anaerobic bacteria and bacterial family enriched in human opportunistic pathogens. Garrett et al[9] reported that Enter-obacteriaceae species act in concert with the gut microbiota to induce colitis. Chen et al[10] also showed that the family Enterobacteriaceae was significantly increased in patients with T2D, which is consistent with our results.
Abnormal metabolism of bile acids is one of the main pathogeneses of cholesterol gallstones,[11] which are mainly adjusted by hepatocytes and the gut microbiota. Host hepatocytes synthesize primary bile acids from cholesterol, after which primary bile acids enter the gastrointestinal tract where they are chemically modified into secondary bile acids. We found that the diastone group had a higher total bile acid level and similar cholesterol level as the stone group. Kakiyama et al[12] reported that higher primary bile acid levels (chenodeoxycholic) were positively correlated with the abundance of Enterobacteriaceae in patients with advanced cirrhosis. These enriched bacterial species in patients with T2D and cholelithiasis may increase the total bile acid levels of the host, leading to gallstone formation.
Thus, maintaining the homeostasis of the gut microbiota and reducing the abundance of Proteobacteria in patients with T2D may help prevent the occurrence of gallstones. However, this study had some limitations. First, 16S rRNA gene amplicon sequencing reveals the composition of microbial community species and cannot directly analyze biological functions related to the species classification. Additionally, we did not perform metabolome analysis to combine different species and metabolic pathways. Further detailed research is required to investigate the mechanism of the enriched bacteria in cholelithiasis.
Funding
This research was supported by grants form the Beijing Natural Science Foundation (Nos. 7192240, 7224351), the National Natural Science Foundation of China (No. 81900386), and the Peking University Shougang Hospital (Nos. SGYYZ201701, SGYYQ201903).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Chen J, Yan L, Ma X, Yuan P, Zhao F, Han Z, Liu J, Wang W, Zhou D, Zhao H, Feng N, Huang D, Hu S, Gu J. Alteration of gut microbiota in type 2 diabetes complicated with cholelithiasis patients. Chin Med J 2022;135:2125–2127. doi: 10.1097/CM9.0000000000002102
Jiajia Chen, Linlin Yan, and Xingfan Ma contributed equally to this work.
Supplemental digital content is available for this article.
References
- 1.Wang L, Gao P, Zhang M, Huang Z, Zhang D, Deng Q, et al. Prevalence and ethnic pattern of diabetes and prediabetes in China in 2013. JAMA 2017; 317:2515–2523. doi: 10.1001/jama.2017.7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song ST, Shi J, Wang XH, Guo YB, Hu PF, Zhu F, et al. Prevalence and risk factors for gallstone disease: a population-based cross-sectional study. J Dig Dis 2020; 21:237–245. doi: 10.1111/1751-2980.12857. [DOI] [PubMed] [Google Scholar]
- 3.Méndez-Sânchez N, Chavez-Tapia NC, Motola-Kuba D, Sanchez-Lara K, Ponciano-Rodriguez G, Baptista H, et al. Metabolic syndrome as a risk factor for gallstone disease. World J Gastroenterol 2005; 11:1653–1657. doi: 10.3748/wjg.v11.i11.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortés VA, Barrera F, Nervi F. Pathophysiological connections between gallstone disease, insulin resistance, and obesity. Obes Rev 2020; 21:e12983–e12991. doi: 10.1111/obr.12983. [DOI] [PubMed] [Google Scholar]
- 5.Aune D, Vatten LJ. Diabetes mellitus and the risk of gallbladder disease: a systematic review and meta-analysis of prospective studies. J Diabetes Complications 2016; 30:368–373. doi: 10.1016/j.jdia-comp.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBiomedicine 2020; 51:1025902–1025910. doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grigor’eva IN, Romanova TI. Gallstone disease and microbiome. Microorganisms 2020; 8:8352–8360. doi: 10.3390/microorganisms8060835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marschall HU, Einarsson C. Gallstone disease. J Intern Med 2007; 261:529–542. doi: 10.1111/j.1365-2796.2007.01783.x. [DOI] [PubMed] [Google Scholar]
- 9.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 2010; 8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Q, Ma X, Li C, Shen Y, Zhu W, Zhang Y, et al. Enteric Phageome alterations in patients with Type 2 diabetes. Front Cell Infect Microbiol 2020; 10:5750842–5750851. doi: 10.3389/fcimb.2020.575084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang DQH, Cohen DE, Carey MC. Biliary lipids and cholesterol gallstone disease. J Lipid Res 2009; 50:S406–S411. doi: 10.1194/jlr.R800075-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 2013; 58:949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.