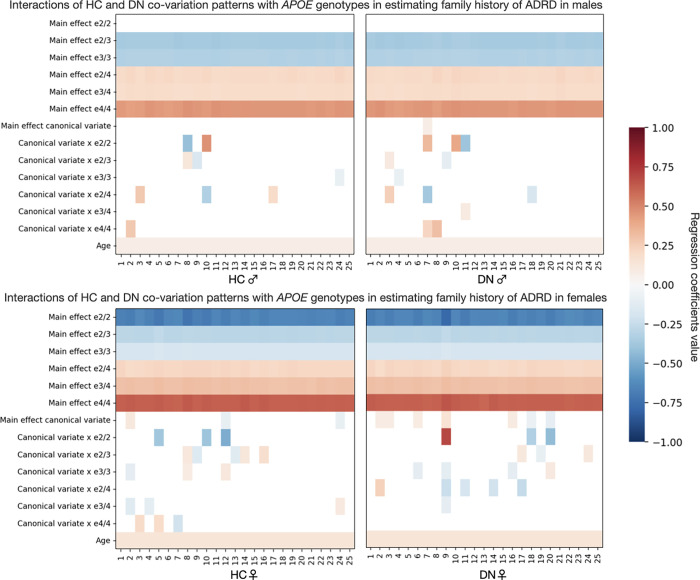
Fig 6. The protectiveness of ɛ2 is sex-dependent and modulated by HC-DN co-variation patterns.
In separate analyses for males and females, we tested whether HC-DN signatures interacted with APOE genotypes in explaining variance in family history of ADRD. Separate analyses were run for males (higher plots) and females (lower plots). Each column on the heat maps represents the coefficients for a single linear regression model. The first 25 columns show the coefficients for HC patterns, whereas the last 25 columns show the coefficients for DN patterns. We assessed the robustness of our findings by comparing each coefficient to empirically built null distributions obtained through permutation testing. Only the coefficients that were statistically different from their respective null distributions 95% of the time are presented. We found that the main effect of APOE ɛ2/2 against ADRD risk was only statistically robust in females. We also showed a spectrum in the opposing effects of ɛ2 and ɛ4 among females, such that ɛ2/4 was associated with a lower increase in ADRD risk than did APOE ɛ3/4, which in turn was associated with lesser risk than ɛ4/4. We further found that the protectiveness of APOE ɛ2/2 interacts with brain structure and can even lead to an increase in ADRD risk among females with a strong expression of mode 9. These interactions profiles suggest that the protectiveness of ɛ2/2 is not only sex-specific but also modulated by HC-DN co-variation expressions. Data underlying this figure can be found at https://github.com/dblabs-mcgill-mila/HCDMNCOV_AD/tree/master/fig_6 (DOI: 10.5281/zenodo.7126809). ADRD, Alzheimer’s disease and related dementia; APOE, Apolipoprotein E; DN, default network; HC, hippocampus.