Figure 6. G protein class-specificity of GN13 and GD20.
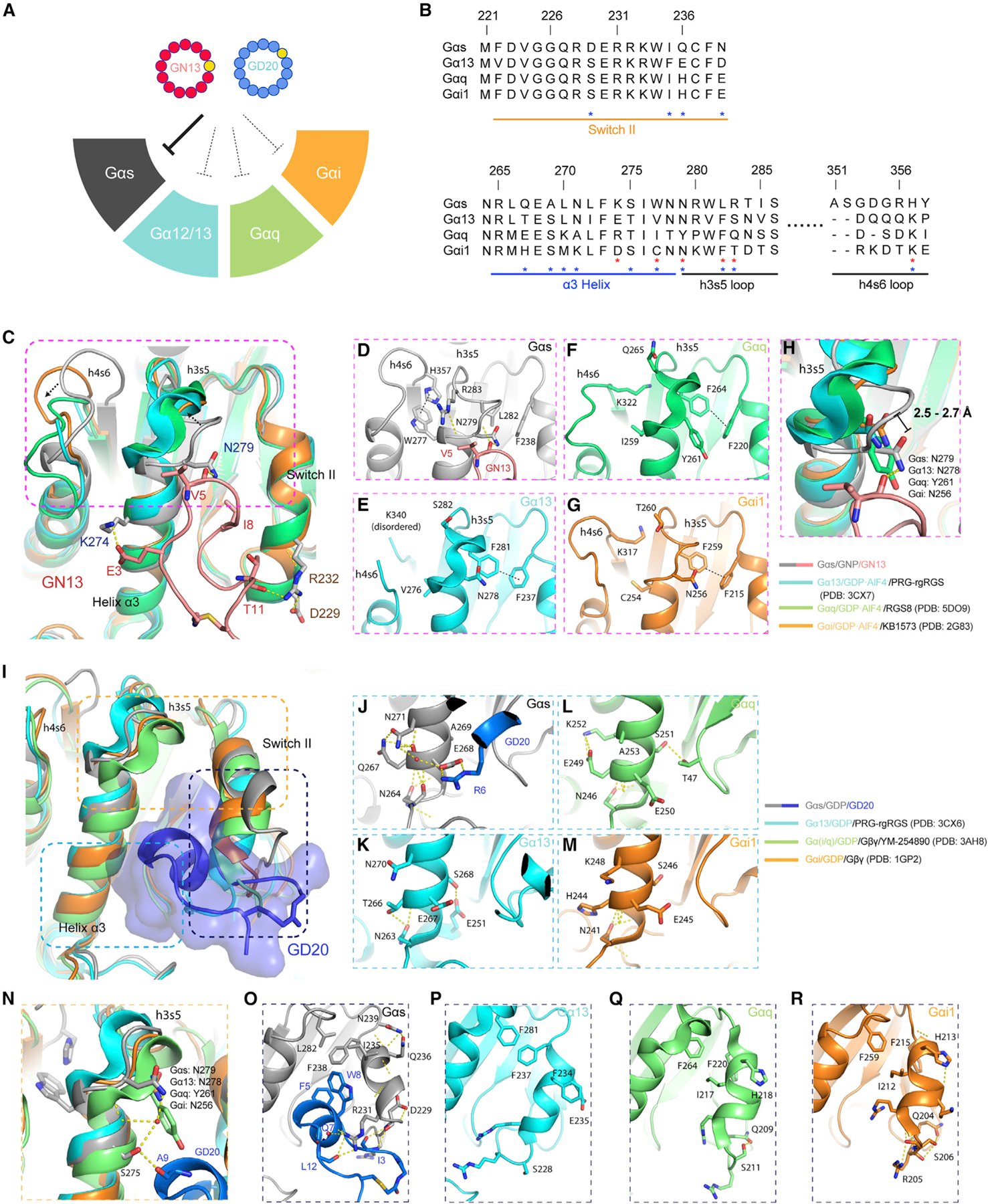
(A) GN13 and GD20 are class-specific Gαs inhibitors.
(B) Sequence alignment of Gα proteins around the cyclic peptide binding site. The residue numbering is based on Gαs. Residues that determine the specificity of GN13 (red) or GD20 (blue) are marked with asterisks.
(C) The active states of Gα13 (cyan, PDB: 3CX7), Gαq (green, PDB: 5DO9), and Gαi (orange, PDB: 2G83) from their complex structures were superimposed on Gαs/GNP in our Gαs (gray)/GN13 (salmon) structure.
(D–G) Structural details of the GN13 binding pocket in different active state Gα proteins. H-bonds are shown as yellow dashed lines. CH/π interactions are shown as black dashed lines.
(H) Close-up view of the critical N279 in Gαs/GNP. Homologous residues in other Gα proteins are labeled with different colors. The distances between the Cα of Gαs N279 and the Cα of other homologous residues are indicated.
(I) The inactive states of Gα13 (cyan, PDB: 3CX6), Gαq (green, PDB: 3AH8), and Gαi (orange, PDB: 1GP2) from their complex structures were superimposed on Gαs/GDP in our Gαs (dark gray)/GD20 (blue) structure.
(J–M) Structural details of the α3 helices in different Gα/GDP proteins.
(N) Close-up view of the critical N279 in Gαs/GDP. Homologous residues in other Gα proteins are labeled with different colors.
(O–R) Structural details of the switch II regions in different Gα/GDP proteins.
See also Figure S6.
