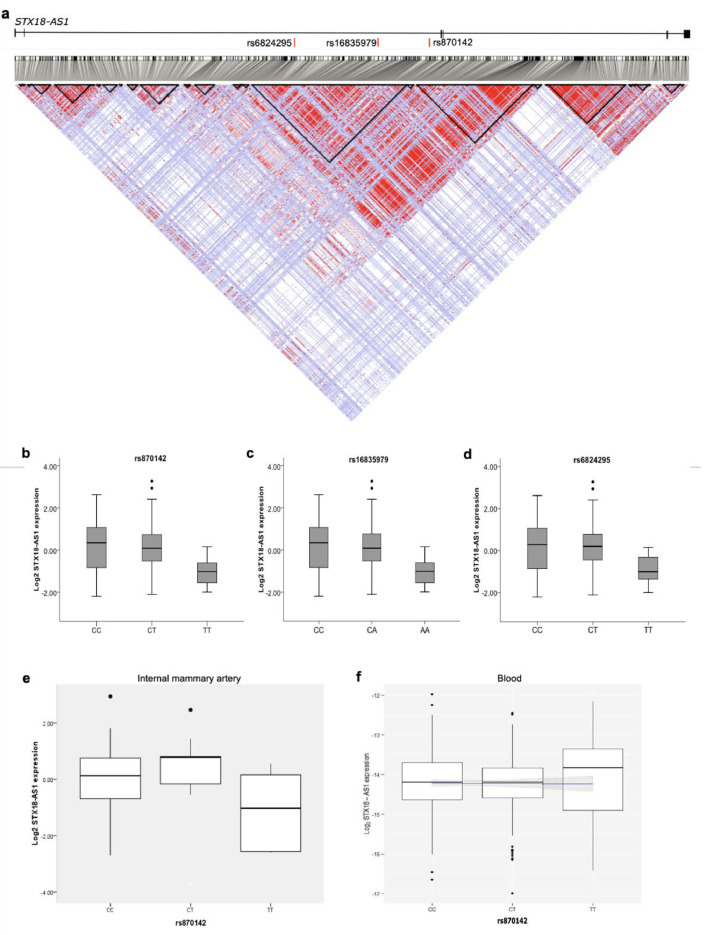
Fig. 2.
Risk SNPs for ASD and eQTL analyses between SNPs and STX18-AS1. a, Linkage disequilibrium (LD) across STX18-AS1 with SNP data from CEU population of 1000 Genomes Project. The LD map was generated with HaploView. The LD of SNP pairs is represented as r2 in white to red (0-1). The locations of the three risk SNPs identified in GWAS are labelled in red bars at the top of the figure. The black triangles represent haplotype blocks (defining with confidence intervals according to Gabriel's method). b-d, eQTL analyses with 108 human heart right atrial appendage (RAA) samples using the qPCR method for three top SNPs identified from GWAS, rs870142 (b), rs16835979(c), and rs6824295(d). The significant association between SNPs and STX18-AS1 transcription was tested using the linear regression model, with P values between 0.038-0.039. e-f, The eQTL analyses of rs870142 and STX18-AS1 transcription in internal mammary artery (IMA) samples (e) and human blood samples (f). The linear regressions are not significant for both IMA and blood eQTL analyses.