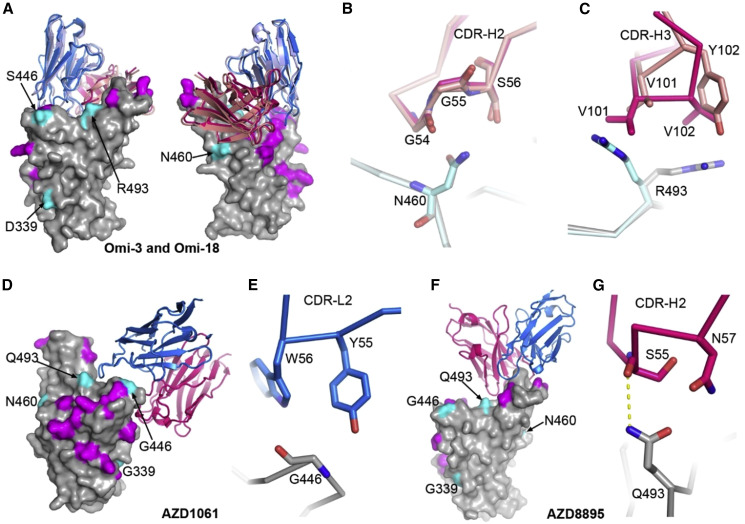
Figure 6.
Interactions between mAbs and BA.2.75 mutation sites
(A) Front and back views of the binding modes of Omi-3 (PDB: 7ZF3) and Omi-18 (PDB: 7ZFC) complexed with Omicron BA.1 RBD by overlapping the RBD. The RBD is shown as a gray surface representation with mutations common to both BA.2 and BA.2.75 colored in magenta, and the four mutations that differ between the two are in cyan. Vhs and Vls are shown as ribbons and colored in red and blue for Omi-3 and light blue and salmon for Omi-18, respectively.
(B) Interactions between N460 of the RBD and CDR-H2 of the Fabs.
(C) Contacts between R493 of the RBD and CDR-H3 of the Fabs.
In (B and C), the RBD associated with Omi-3 is in gray and Omi-18 in cyan, and the colors of the Fabs are as in (A).
(D and E) AZD1061 bound with the ancestral SARS-CoV-2 RBD (PDB: 7L7E) (D) and contacts between G446 of the RBD and CDR-L2 of the Fab (E).
(F and G) AZD8895 bound with the ancestral SARS-CoV-2 spike RBD (PDB: 7L7E) (F) and contacts between Q493 of the RBD and CDR-H2 of the Fab (G).
In (D–F), the RBD is drawn and colored as in (A), and heavy chain is in red and light chain in blue.