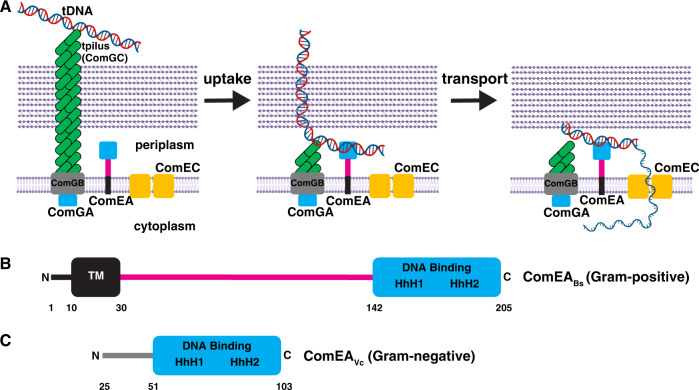
Fig. 1. Existing model of genetic transformation in Gram-positive bacteria, and ComEA domain architecture in Gram-positive and Gram-negative bacteria.
A The tpilus is composed of ComGC, whose assembly requires the ATPase ComGA. The tpilus is believed to be anchored to the membrane protein ComGB. The tpilus binds weakly to DNA and retracts to pull it into the periplasm. Here, the DNA encounters ComEA, which stabilizes binding to the cell and propels continued uptake of the DNA. ComEC is proposed to degrade one strand of DNA and provide the channel for transport of DNA into the cytoplasm. B Domain architecture of ComEA from a representative Gram-positive bacterium, B. subtilis. C Domain architecture of ComEA from a representative Gram-negative bacterium, V. cholerae. Residues 1–24 are not shown in order to highlight the fact that they comprise a predicted secretion signal that is cleaved to generate mature ComEAVc, which diffuses freely in the periplasm. TM, predicted transmembrane region. The magenta line denotes a region of unknown function, which is addressed in this study. HhH, helix-hairpin-helix motifs. Residue numbering corresponds to ComEABs or ComEAVc. Elements of the figure were created with BioRender.com.