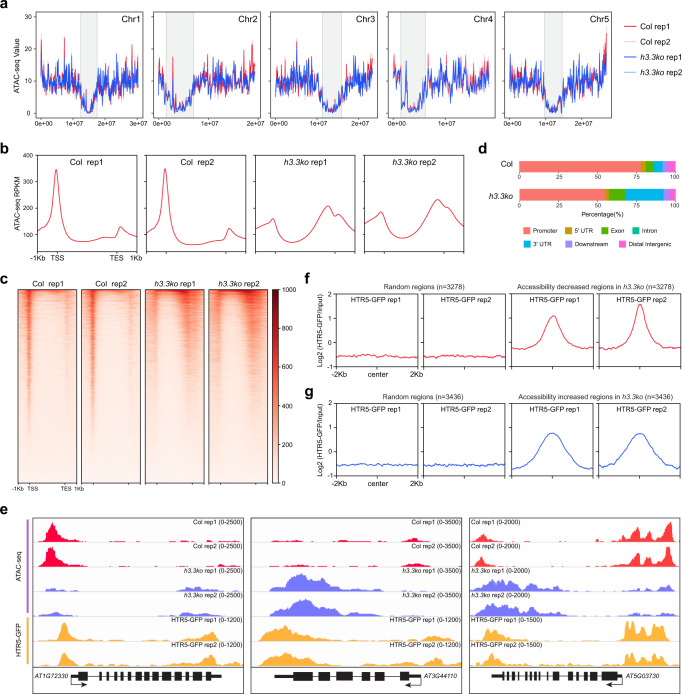
Fig. 4. Chromatin accessibility changes in mature h3.3ko seeds.
a ATAC-seq signals in Col and h3.3ko mature seeds over Arabidopsis chromosomes. Signals were calculated in 100 kb bins. Pericentromeric heterochromatin regions are indicated with gray shading. b Metaplot of ATAC-seq signals in Col and h3.3ko mature seeds over all genes. c Heatmap of ATAC-seq signals in Col and h3.3ko mature seeds over all genes. d Genomic distribution of accessible regions in Col and h3.3ko mature seeds. e Genome browser view of ATAC-seq and HTR5-GFP ChIP-seq signals in Col and h3.3ko mature seeds over a few representative loci. AT1G72330: a gene mainly lost accessibility at the 5′ end/promoter region in h3.3ko, AT3G44110: a gene mainly gained accessibility around the 3′ end/genic region in h3.3ko, AT5G03730: a gene with both accessibility increased and decreased regions in h3.3ko. f, g Metaplot of HTR5-GFP ChIP-seq signals in mature seeds over accessibility significantly decreased (f) and increased (g) regions in mature h3.3ko seeds compared with Col. HTR5-GFP ChIP-seq signals over randomly selected regions are served as control.