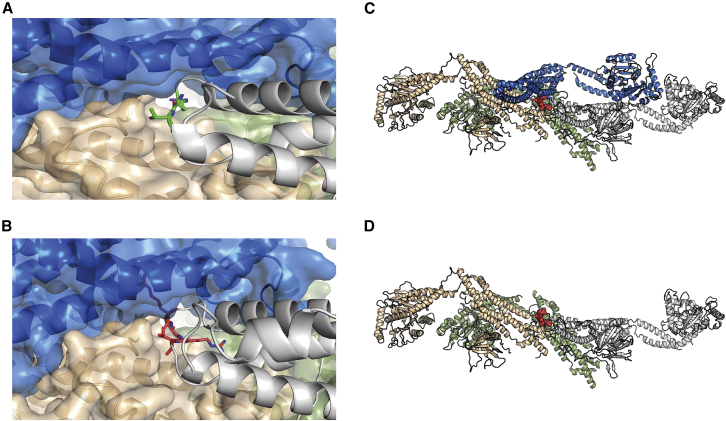
Figure 4.
Structural modeling of the effects of the c.1197−8G>A (GenBank: NM_001288739.1) variant
(A) Mutation site in dynamin-1 corresponding to c.1197−8G>A (GenBank: NM_001288739.1). Green, two residues neighboring the CR insertion.
(B) Model of the CR insertion (pink). Given the lack of space to accommodate, the new residues steric clashes would be created, disrupting the interactions to the neighboring protomers.
(C and D) Tetrameric organization of DNM1 (PDB ID: 5A3F). The tetramer is formed by two sets of parallel protomers facing in the same direction. Gray and green and blue and orange. When a mutant protomer containing the CR insertion (bottom: gray chain) forms part of the complex, the corresponding parallel protomer (blue) can no longer bind as a result of steric hindrance.