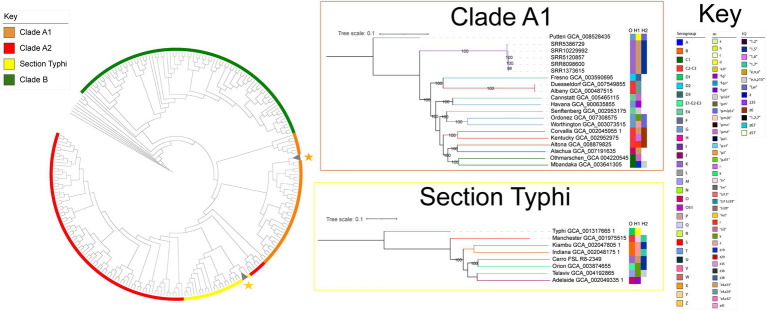
Figure 1.
S. Cerro is polyphyletic and the majority of isolates cluster within the Salmonella section Typhi lineage. Maximum likelihood phylogenetic tree constructed from 12,238 core SNPs for representative isolates of S. Cerro and isolates representing 235 S. enterica subsp. enterica serovars and five Salmonella subspecies (Gaballa et al., 2021); branch lengths are not shown. Clade designations shown are based on those defined previously (Worley et al., 2018). Salmonella enterica subsp. arizonae was used as an outgroup to root the phylogeny. Maximum likelihood trees of the sub-clades in which S. Cerro isolates cluster are also shown. The Clade A1 tree was constructed from 60,135 core SNPs and the section Typhi tree was constructed from 46,278 core SNPs. Phylogenies are rooted by outgroup (the clade A1 phylogeny uses S. Putten GCA_008528435 as an outgroup, the section Typhi phylogeny uses S. Typhi GCA_001317665.1 as an outgroup; bootstrap values represent the average of 1,000 bootstrap repetitions).